数据总目录:
/home/bavon/datasets/wsi/hsil
/home/bavon/datasets/wsi/lsil
1 规整文件命名以及xml拷贝
data_prepare.py 的 align_xml_svs 方法
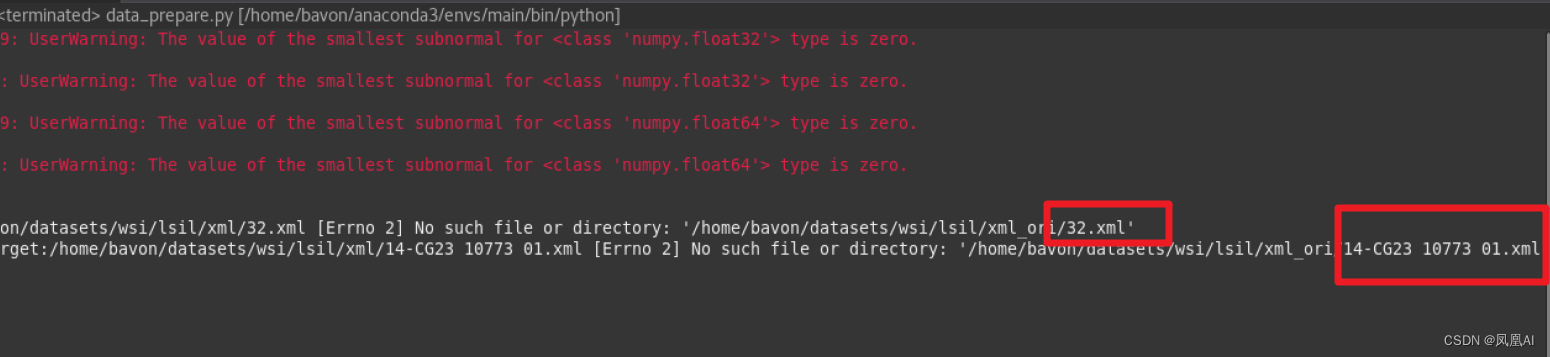
if __name__ == '__main__': file_path = "/home/bavon/datasets/wsi/lsil"# align_xml_svs(file_path) # build_data_csv(file_path)# crop_with_annotation(file_path)# build_annotation_patches(file_path)# aug_annotation_patches(file_path)# filter_patches_exclude_anno(file_path)# build_normal_patches_image(file_path)用到哪个方法将其进行注释打开
def align_xml_svs(file_path):"""Solving the problem of inconsistent naming between SVS and XML"""wsi_path = file_path + "/data"ori_xml_path = file_path + "/xml_ori"target_xml_path = file_path + "/xml"for wsi_file in os.listdir(wsi_path):if not wsi_file.endswith(".svs"):continue single_name = wsi_file.split(".")[0]if "-" in single_name and False:xml_single_name = single_name.split("-")[0]else:xml_single_name = single_namexml_single_name = xml_single_name + ".xml"ori_xml_file = os.path.join(ori_xml_path,xml_single_name)tar_xml_file = os.path.join(target_xml_path,single_name + ".xml")try:copyfile(ori_xml_file,tar_xml_file)except Exception as e:print("copyfile fail,source:{} and target:{}".format(ori_xml_file,tar_xml_file),e)
2 生成normal切片(默认level1)
create_patches_fp.py
输入目录 data 输出目录 patches_level1
--source /home/bavon/datasets/wsi/lsil --save_dir /home/bavon/datasets/wsi/lsil --step_size 64 --patch_size 64 --seg --patch --stitch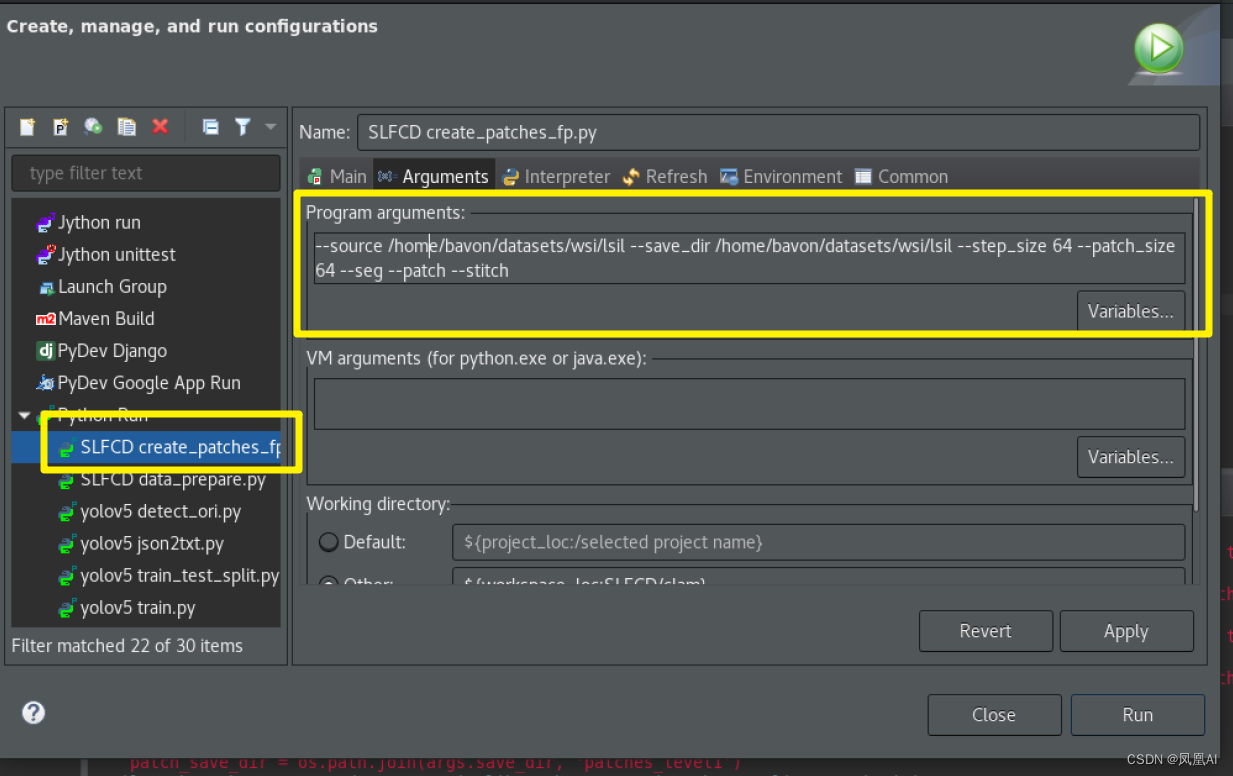
# internal imports
from wsi_core.WholeSlideImage import WholeSlideImage
from wsi_core.wsi_utils import StitchCoords
from wsi_core.batch_process_utils import initialize_df
# other imports
import os
import numpy as np
import time
import argparse
import pdb
import pandas as pddef stitching(file_path, wsi_object, downscale = 64):start = time.time()heatmap = StitchCoords(file_path, wsi_object, downscale=downscale, bg_color=(0,0,0), alpha=-1, draw_grid=False)total_time = time.time() - startreturn heatmap, total_timedef segment(WSI_object, seg_params = None, filter_params = None, mask_file = None):### Start Seg Timerstart_time = time.time()# Use segmentation fileif mask_file is not None:WSI_object.initSegmentation(mask_file)# Segment else:WSI_object.segmentTissue(**seg_params, filter_params=filter_params)### Stop Seg Timersseg_time_elapsed = time.time() - start_time return WSI_object, seg_time_elapseddef patching(WSI_object, **kwargs):### Start Patch Timerstart_time = time.time()# Patchfile_path = WSI_object.process_contours(**kwargs)### Stop Patch Timerpatch_time_elapsed = time.time() - start_timereturn file_path, patch_time_elapseddef seg_and_patch(source, save_dir, patch_save_dir, mask_save_dir, stitch_save_dir, patch_size = 256, step_size = 256, seg_params = {'seg_level': -1, 'sthresh': 8, 'mthresh': 7, 'close': 4, 'use_otsu': False,'keep_ids': 'none', 'exclude_ids': 'none'},filter_params = {'a_t':100, 'a_h': 16, 'max_n_holes':8}, vis_params = {'vis_level': -1, 'line_thickness': 500},patch_params = {'use_padding': True, 'contour_fn': 'four_pt'},patch_level = 0,use_default_params = False, seg = False, save_mask = True, stitch= False, patch = False, auto_skip=True, process_list = None):wsi_source = os.path.join(source,"data")slides = sorted(os.listdir(wsi_source))slides = [slide for slide in slides if os.path.isfile(os.path.join(wsi_source, slide))]if process_list is None:df = initialize_df(slides, seg_params, filter_params, vis_params, patch_params)else:df = pd.read_csv(process_list)df = initialize_df(df, seg_params, filter_params, vis_params, patch_params)mask = df['process'] == 1process_stack = df[mask]total = len(process_stack)legacy_support = 'a' in df.keys()if legacy_support:print('detected legacy segmentation csv file, legacy support enabled')df = df.assign(**{'a_t': np.full((len(df)), int(filter_params['a_t']), dtype=np.uint32),'a_h': np.full((len(df)), int(filter_params['a_h']), dtype=np.uint32),'max_n_holes': np.full((len(df)), int(filter_params['max_n_holes']), dtype=np.uint32),'line_thickness': np.full((len(df)), int(vis_params['line_thickness']), dtype=np.uint32),'contour_fn': np.full((len(df)), patch_params['contour_fn'])})seg_times = 0.patch_times = 0.stitch_times = 0.for i in range(total):df.to_csv(os.path.join(save_dir, 'process_list_autogen.csv'), index=False)idx = process_stack.index[i]slide = process_stack.loc[idx, 'slide_id']if not slide.endswith(".svs"):continueprint("\n\nprogress: {:.2f}, {}/{}".format(i/total, i, total))print('processing {}'.format(slide))df.loc[idx, 'process'] = 0slide_id, _ = os.path.splitext(slide)if auto_skip and os.path.isfile(os.path.join(patch_save_dir, slide_id + '.h5')):print('{} already exist in destination location, skipped'.format(slide_id))df.loc[idx, 'status'] = 'already_exist'continue# Inialize WSIfull_path = os.path.join(source, "data",slide)xml_file = slide.replace(".svs",".xml")xml_path = os.path.join(source,"xml", xml_file)tumor_mask_file = slide.replace(".svs",".npy")tumor_mask_path = os.path.join(source,"tumor_mask", tumor_mask_file)if not os.path.exists(xml_path):df.loc[idx, 'status'] = 'failed_seg'continueWSI_object = WholeSlideImage(full_path)WSI_object.initXML(xml_path)
# WSI_object.initMask(tumor_mask_path)if use_default_params:current_vis_params = vis_params.copy()current_filter_params = filter_params.copy()current_seg_params = seg_params.copy()current_patch_params = patch_params.copy()else:current_vis_params = {}current_filter_params = {}current_seg_params = {}current_patch_params = {}for key in vis_params.keys():if legacy_support and key == 'vis_level':df.loc[idx, key] = -1current_vis_params.update({key: df.loc[idx, key]})for key in filter_params.keys():if legacy_support and key == 'a_t':old_area = df.loc[idx, 'a']seg_level = df.loc[idx, 'seg_level']scale = WSI_object.level_downsamples[seg_level]adjusted_area = int(old_area * (scale[0] * scale[1]) / (512 * 512))current_filter_params.update({key: adjusted_area})df.loc[idx, key] = adjusted_areacurrent_filter_params.update({key: df.loc[idx, key]})for key in seg_params.keys():if legacy_support and key == 'seg_level':df.loc[idx, key] = -1current_seg_params.update({key: df.loc[idx, key]})for key in patch_params.keys():current_patch_params.update({key: df.loc[idx, key]})if current_vis_params['vis_level'] < 0:if len(WSI_object.level_dim) == 1:current_vis_params['vis_level'] = 0else: wsi = WSI_object.getOpenSlide()best_level = wsi.get_best_level_for_downsample(64)current_vis_params['vis_level'] = best_levelif current_seg_params['seg_level'] < 0:if len(WSI_object.level_dim) == 1:current_seg_params['seg_level'] = 0else:wsi = WSI_object.getOpenSlide()best_level = wsi.get_best_level_for_downsample(64)current_seg_params['seg_level'] = best_levelkeep_ids = str(current_seg_params['keep_ids'])if keep_ids != 'none' and len(keep_ids) > 0:str_ids = current_seg_params['keep_ids']current_seg_params['keep_ids'] = np.array(str_ids.split(',')).astype(int)else:current_seg_params['keep_ids'] = []exclude_ids = str(current_seg_params['exclude_ids'])if exclude_ids != 'none' and len(exclude_ids) > 0:str_ids = current_seg_params['exclude_ids']current_seg_params['exclude_ids'] = np.array(str_ids.split(',')).astype(int)else:current_seg_params['exclude_ids'] = []w, h = WSI_object.level_dim[current_seg_params['seg_level']] if w * h > 1e8:print('level_dim {} x {} is likely too large for successful segmentation, aborting'.format(w, h))df.loc[idx, 'status'] = 'failed_seg'continuedf.loc[idx, 'vis_level'] = current_vis_params['vis_level']df.loc[idx, 'seg_level'] = current_seg_params['seg_level']seg_time_elapsed = -1if seg:WSI_object, seg_time_elapsed = segment(WSI_object, current_seg_params, current_filter_params) if save_mask:mask = WSI_object.visWSI(**current_vis_params)mask_path = os.path.join(mask_save_dir, slide_id+'.jpg')mask.save(mask_path)patch_time_elapsed = -1 # Default timeif patch:current_patch_params.update({'patch_level': patch_level, 'patch_size': patch_size, 'step_size': step_size, 'save_path': patch_save_dir})file_path, patch_time_elapsed = patching(WSI_object = WSI_object, **current_patch_params,)stitch_time_elapsed = -1if stitch:file_path = os.path.join(patch_save_dir, slide_id+'.h5')if os.path.isfile(file_path):heatmap, stitch_time_elapsed = stitching(file_path, WSI_object, downscale=64)stitch_path = os.path.join(stitch_save_dir, slide_id+'.jpg')heatmap.save(stitch_path)print("segmentation took {} seconds".format(seg_time_elapsed))print("patching took {} seconds".format(patch_time_elapsed))print("stitching took {} seconds".format(stitch_time_elapsed))df.loc[idx, 'status'] = 'processed'seg_times += seg_time_elapsedpatch_times += patch_time_elapsedstitch_times += stitch_time_elapsedseg_times /= totalpatch_times /= totalstitch_times /= totaldf = df[df["status"]!="failed_seg"]df.to_csv(os.path.join(save_dir, 'process_list_autogen.csv'), index=False)print("average segmentation time in s per slide: {}".format(seg_times))print("average patching time in s per slide: {}".format(patch_times))print("average stiching time in s per slide: {}".format(stitch_times))return seg_times, patch_timesparser = argparse.ArgumentParser(description='seg and patch')
parser.add_argument('--source', type = str,help='path to folder containing raw wsi image files')
parser.add_argument('--step_size', type = int, default=256,help='step_size')
parser.add_argument('--patch_size', type = int, default=256,help='patch_size')
parser.add_argument('--patch', default=False, action='store_true')
parser.add_argument('--seg', default=False, action='store_true')
parser.add_argument('--stitch', default=False, action='store_true')
parser.add_argument('--no_auto_skip', default=True, action='store_false')
parser.add_argument('--save_dir', type = str,help='directory to save processed data')
parser.add_argument('--preset', default=None, type=str,help='predefined profile of default segmentation and filter parameters (.csv)')
parser.add_argument('--patch_level', type=int, default=1, help='downsample level at which to patch')
parser.add_argument('--process_list', type = str, default=None,help='name of list of images to process with parameters (.csv)')if __name__ == '__main__':args = parser.parse_args()patch_save_dir = os.path.join(args.save_dir, 'patches_level1')mask_save_dir = os.path.join(args.save_dir, 'masks')stitch_save_dir = os.path.join(args.save_dir, 'stitches')if args.process_list:process_list = os.path.join(args.save_dir, args.process_list)else:process_list = Noneprint('source: ', args.source)print('patch_save_dir: ', patch_save_dir)print('mask_save_dir: ', mask_save_dir)print('stitch_save_dir: ', stitch_save_dir)directories = {'source': args.source, 'save_dir': args.save_dir,'patch_save_dir': patch_save_dir, 'mask_save_dir' : mask_save_dir, 'stitch_save_dir': stitch_save_dir} for key, val in directories.items():print("{} : {}".format(key, val))if key not in ['source']:os.makedirs(val, exist_ok=True)seg_params = {'seg_level': -1, 'sthresh': 8, 'mthresh': 7, 'close': 4, 'use_otsu': False,'keep_ids': 'none', 'exclude_ids': 'none'}filter_params = {'a_t':100, 'a_h': 16, 'max_n_holes':8}vis_params = {'vis_level': -1, 'line_thickness': 250}patch_params = {'use_padding': True, 'contour_fn': 'four_pt'}if args.preset:preset_df = pd.read_csv(os.path.join('presets', args.preset))for key in seg_params.keys():seg_params[key] = preset_df.loc[0, key]for key in filter_params.keys():filter_params[key] = preset_df.loc[0, key]for key in vis_params.keys():vis_params[key] = preset_df.loc[0, key]for key in patch_params.keys():patch_params[key] = preset_df.loc[0, key]parameters = {'seg_params': seg_params,'filter_params': filter_params,'patch_params': patch_params,'vis_params': vis_params}print(parameters)seg_times, patch_times = seg_and_patch(**directories, **parameters,patch_size = args.patch_size, step_size=args.step_size, seg = args.seg, use_default_params=False, save_mask = True, stitch= args.stitch,patch_level=args.patch_level, patch = args.patch,process_list = process_list, auto_skip=args.no_auto_skip)
3 xml标注文件转json
utexml2json2.py
Ctrl+Shift+R 找找
import sys
import os
import argparse
import loggingsys.path.append(os.path.dirname(os.path.abspath(__file__)) + '/../../../')from camelyon16.data.annotation import UteFormatterparser = argparse.ArgumentParser(description='Convert Camelyon16 xml format to''internal json format')def run(args):# file_path = "/home/bavon/datasets/wsi/hsil"file_path = "/home/bavon/datasets/wsi/lsil"xml_path = os.path.join(file_path,"xml")json_path = os.path.join(file_path,"json")for file in os.listdir(xml_path):json_file = file.replace("xml", "json") json_file_path = os.path.join(json_path,json_file)xml_file_path = os.path.join(xml_path,file)UteFormatter().xml2json(xml_file_path, json_file_path)def main():logging.basicConfig(level=logging.INFO)args = parser.parse_args()run(args)if __name__ == '__main__':main()
4 tumor_mask.py
输出目录 tumor_mask_level1
Label_Dict = [{"code":1,"group_code":"D","desc":"CIN 2"},{"code":2,"group_code":"E","desc":"CIN 3"},{"code":3,"group_code":"F","desc":"CIN 2 to 3"},]将其进行修改,修改后为
Label_Dict = [{"code":1,"group_code":"D","desc":"CIN 2"},{"code":2,"group_code":"E","desc":"CIN 3"},{"code":3,"group_code":"F","desc":"CIN 2 to 3"},{"code":4,"group_code":"A","desc":"Large hollowed-out cells, transparent"},{"code":5,"group_code":"B","desc":"The nucleus is deeply stained, small, and heterotypic"},{"code":6,"group_code":"C","desc":"Small hollowed-out cells, transparent"},]def get_label_with_group_code(group_code):for item in Label_Dict:if group_code==item["group_code"]:return itemdef get_label_cate():cate = [0,1,2,3,4,5,6]# cate = [0,1]return cate def get_tumor_label_cate():return [1,2,3,4,5,6]import os
import sys
import logging
import argparseimport numpy as np
import openslide
import cv2
import jsonfrom utils.constance import get_label_with_group_codesys.path.append(os.path.dirname(os.path.abspath(__file__)) + '/../../')parser = argparse.ArgumentParser(description='Get tumor mask of tumor-WSI and ''save it in npy format')
parser.add_argument('wsi_path', default=None, metavar='WSI_PATH', type=str,help='Path to the WSI file')
parser.add_argument('json_path', default=None, metavar='JSON_PATH', type=str,help='Path to the JSON file')
parser.add_argument('npy_path', default=None, metavar='NPY_PATH', type=str,help='Path to the output npy mask file')
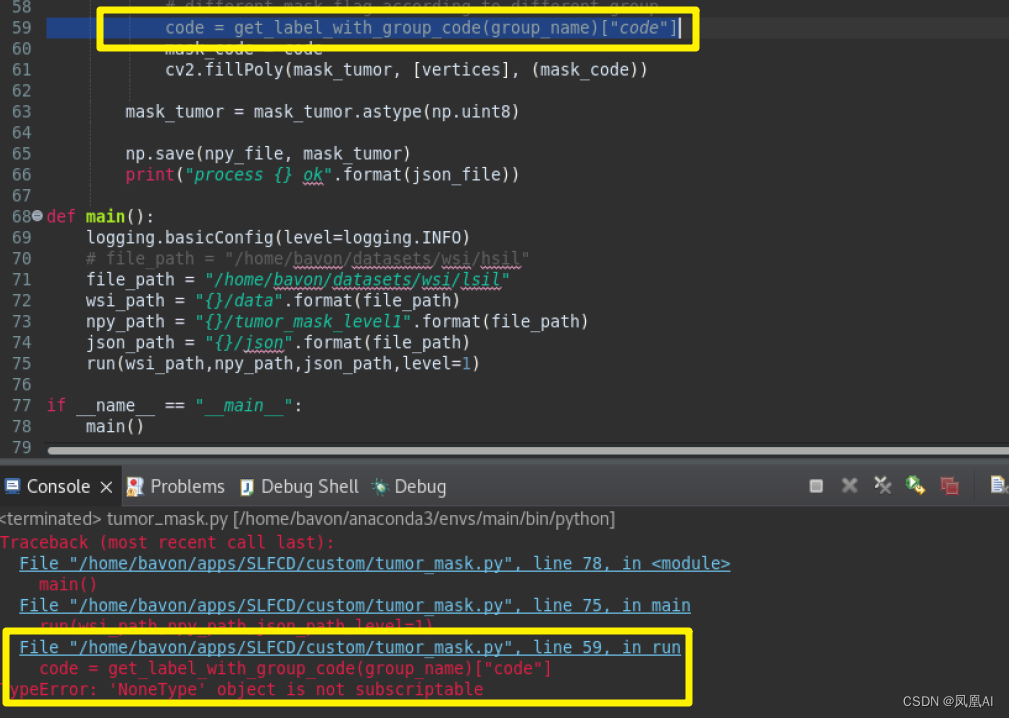
parser.add_argument('--level', default=6, type=int, help='at which WSI level'' to obtain the mask, default 6')def run(wsi_path,npy_path,json_path,level=0):for json_file in os.listdir(json_path):json_file_path = os.path.join(json_path,json_file)single_name = json_file.split(".")[0]npy_file = os.path.join(npy_path,single_name+".npy")wsi_file_path = os.path.join(wsi_path,single_name+".svs")slide = openslide.OpenSlide(wsi_file_path)if len(slide.level_dimensions)<=level:print("no level for {},ignore:".format(wsi_file_path))continue w, h = slide.level_dimensions[level]mask_tumor = np.zeros((h, w)) # the init mask, and all the value is 0# get the factor of level * e.g. level 6 is 2^6factor = slide.level_downsamples[level]try:with open(json_file_path) as f:dicts = json.load(f)except Exception as e:print("open json file fail,ignore:{}".format(json_file_path))continuetumor_polygons = dicts['positive']for tumor_polygon in tumor_polygons:# plot a polygonname = tumor_polygon["name"]group_name = tumor_polygon["group_name"]vertices = np.array(tumor_polygon["vertices"]) / factorvertices = vertices.astype(np.int32)# different mask flag according to different group code = get_label_with_group_code(group_name)["code"]mask_code = codecv2.fillPoly(mask_tumor, [vertices], (mask_code))mask_tumor = mask_tumor.astype(np.uint8)np.save(npy_file, mask_tumor)print("process {} ok".format(json_file))def main():logging.basicConfig(level=logging.INFO)file_path = "/home/bavon/datasets/wsi/lsil"wsi_path = "{}/data".format(file_path) npy_path = "{}/tumor_mask_level1".format(file_path) json_path = "{}/json".format(file_path) run(wsi_path,npy_path,json_path,level=1)if __name__ == "__main__":main()
5 生成训练和测试数据对照表 csv
data_prepare.py 的 build_data_csv 方法
def build_data_csv(file_path,split_rate=0.7):"""build train and valid list to csv"""wsi_path = file_path + "/data"xml_path = file_path + "/xml"total_file_number = len(os.listdir(xml_path))train_number = int(total_file_number * split_rate)train_file_path = file_path + "/train.csv"valid_file_path = file_path + "/valid.csv"list_train = []list_valid = []for i,xml_file in enumerate(os.listdir(xml_path)):single_name = xml_file.split(".")[0]wsi_file = single_name + ".svs"if i < train_number:list_train.append([wsi_file,1])else:list_valid.append([wsi_file,1])train_df = pd.DataFrame(np.array(list_train),columns=['slide_id','label'])valid_df = pd.DataFrame(np.array(list_valid),columns=['slide_id','label'])train_df.to_csv(train_file_path,index=False,sep=',')valid_df.to_csv(valid_file_path,index=False,sep=',')
6 生成标注对应的图片patch
data_prepare.py 的 crop_with_annotation 以及 build_annotation_patches 方法
输出目录 tumor_patch_img
def crop_with_annotation(file_path,level=1):"""Crop image from WSI refer to annotation"""crop_img_path = file_path + "/crop_img"patch_path = file_path + "/patches_level{}".format(level)wsi_path = file_path + "/data"json_path = file_path + "/json"total_file_number = len(os.listdir(json_path))for i,json_file in enumerate(os.listdir(json_path)):json_file_path = os.path.join(json_path,json_file) single_name = json_file.split(".")[0] wsi_file = os.path.join(wsi_path,single_name + ".svs") wsi = openslide.open_slide(wsi_file) scale = wsi.level_downsamples[level]with open(json_file_path, 'r') as jf:anno_data = json.load(jf)# Convert irregular annotations to rectanglesregion_data = []label_data = []for i,anno_item in enumerate(anno_data["positive"]):vertices = np.array(anno_item["vertices"])group_name = anno_item["group_name"]label = get_label_with_group_code(group_name)['code']label_data.append(label)x_min = vertices[:,0].min()x_max = vertices[:,0].max()y_min = vertices[:,1].min()y_max = vertices[:,1].max()region_size = (int((x_max - x_min)/scale),int((y_max-y_min)/scale))xywh = [x_min,y_min,region_size[0],region_size[1]]region_data.append(xywh)# crop_img = np.array(wsi.read_region((x_min,y_min), level, region_size).convert("RGB"))# crop_img = cv2.cvtColor(crop_img,cv2.COLOR_RGB2BGR) # img_file_name = "{}_{}|{}.jpg".format(single_name,i,label)# img_file_path = os.path.join(crop_img_path,img_file_name)# cv2.imwrite(img_file_path,crop_img)# print("save image:{}".format(img_file_name))# Write region data to H5patch_file_path = os.path.join(patch_path,single_name+".h5") with h5py.File(patch_file_path, "a") as f:if "crop_region" in f:del f["crop_region"]f.create_dataset('crop_region', data=np.array(region_data)) f['crop_region'].attrs['label_data'] = label_datadef build_annotation_patches(file_path,level=1,patch_size=64):"""Load and build positive annotation data"""patch_path = file_path + "/patches_level{}".format(level)wsi_path = file_path + "/data"for patch_file in os.listdir(patch_path):file_name = patch_file.split(".")[0]# if file_name!="9-CG23_12974_12":# continuepatch_file_path = os.path.join(patch_path,patch_file)wsi_file_path = os.path.join(wsi_path,file_name+".svs")wsi = openslide.open_slide(wsi_file_path)scale = wsi.level_downsamples[level]mask_path = os.path.join(file_path,"tumor_mask_level{}".format(level))npy_file = os.path.join(mask_path,file_name+".npy") mask_data = np.load(npy_file)with h5py.File(patch_file_path, "a") as f:print("crop_region for:{}".format(patch_file_path))crop_region = f['crop_region'][:]label_data = f['crop_region'].attrs['label_data'] patches = []patches_length = 0db_keys = []for i in range(len(label_data)):region = crop_region[i]label = label_data[i]# Patch for every annotation images,Build patches coordinate data list patch_data = patch_anno_img(region,mask_data=mask_data,patch_size=patch_size,scale=scale,file_path=file_path,file_name=file_name,label=label,index=i,level=level,wsi=wsi) if patch_data is None:# viz_crop_patch(file_path,file_name,region,None) patch_data = np.array([])patches_length += patch_data.shape[0]db_key = "anno_patches_data_{}".format(i)if db_key in f:del f[db_key]f.create_dataset(db_key, data=patch_data)db_keys.append(db_key)if "annotations" in f:del f["annotations"]# annotation summarizef.create_dataset("annotations", data=db_keys) # Record total length and labelf["annotations"].attrs['patches_length'] = patches_length f["annotations"].attrs['label_data'] = label_data print("patch {} ok".format(file_name))
7 生成未标注区域对应的图片patch
data_prepare.py 的 build_normal_patches_image 方法
输出目录 tumor_patch_img
def build_normal_patches_image(file_path,level=1,patch_size=64):"""Build images of normal region in wsi"""patch_path = file_path + "/patches_level{}".format(level)wsi_path = file_path + "/data"for patch_file in os.listdir(patch_path):file_name = patch_file.split(".")[0]patch_file_path = os.path.join(patch_path,patch_file)wsi_file_path = os.path.join(wsi_path,file_name+".svs")wsi = openslide.open_slide(wsi_file_path)scale = wsi.level_downsamples[level]mask_path = os.path.join(file_path,"tumor_mask_level{}".format(level))npy_file = os.path.join(mask_path,file_name+".npy") mask_data = np.load(npy_file)save_path = os.path.join(file_path,"tumor_patch_img/0",file_name)if not os.path.exists(save_path):os.mkdir(save_path)print("process file:{}".format(patch_file_path))with h5py.File(patch_file_path, "a") as f:if not "coords" in f:print("coords not in:{}".format(file_name)) continue coords = f['coords'][:]for idx,coord in enumerate(coords):# Ignore annotation patches dataif judge_patch_anno(coord,mask_data=mask_data,scale=scale,patch_size=patch_size):continuecrop_img = np.array(wsi.read_region(coord, level, (patch_size,patch_size)).convert("RGB"))crop_img = cv2.cvtColor(crop_img,cv2.COLOR_RGB2BGR) save_file_path = os.path.join(save_path,"{}.jpg".format(idx))cv2.imwrite(save_file_path,crop_img)print("write image ok:{}".format(file_name))8 训练 train_with_clamdata.py
import sys
import os
import shutil
import argparse
import logging
import json
import time
from argparse import Namespace
import pandas as pd
import torch
from torch.utils.data import DataLoader
from torch.autograd import Variable
from torch.nn import BCEWithLogitsLoss, DataParallel
from torch.optim import SGD
from torchvision import models
from torch import nn
from tensorboardX import SummaryWriterimport torch
import torch.nn as nn
# import torch.nn.LSTM
import torchvision
import torchvision.transforms as transforms
import torch.nn.functional as F
import matplotlib.pyplot as plt
import pytorch_lightning as pl
from pytorch_lightning import loggers as pl_loggers
from pytorch_lightning import Trainer
from pytorch_lightning.callbacks import ModelCheckpoint
from pytorch_lightning.trainer.states import RunningStage
import numpy as np
from clam.datasets.dataset_h5 import Dataset_All_Bags
# from clam.datasets.dataset_combine import Whole_Slide_Bag_COMBINE
from clam.datasets.dataset_combine_together import Whole_Slide_Bag_COMBINE_togeter
from clam.utils.utils import print_network, collate_features
import typessys.path.append(os.path.dirname(os.path.abspath(__file__)) + '/../../')from camelyon16.data.image_producer import ImageDataset
from utils.constance import get_label_cateparser = argparse.ArgumentParser(description='Train model')
parser.add_argument('cnn_path', default=None, metavar='CNN_PATH', type=str,help='Path to the config file in json format')
parser.add_argument('save_path', default=None, metavar='SAVE_PATH', type=str,help='Path to the saved models')
parser.add_argument('--num_workers', default=2, type=int, help='number of'' workers for each data loader, default 2.')
parser.add_argument('--device_ids', default='0', type=str, help='comma'' separated indices of GPU to use, e.g. 0,1 for using GPU_0'' and GPU_1, default 0.')device = 'cuda:0' # torch.device('cuda:0')
os.environ["CUDA_VISIBLE_DEVICES"] = '0'
# device = torch.device('cpu')from utils.vis import vis_data,visdom_data
from visdom import Visdomviz_tumor_train = Visdom(env="tumor_train", port=8098)
viz_tumor_valid = Visdom(env="tumor_valid", port=8098)
viz_normal_train = Visdom(env="normal_train", port=8098)
viz_normal_valid = Visdom(env="normal_valid", port=8098)def chose_model(model_name):if model_name == 'resnet18':model = models.resnet18(pretrained=False)elif model_name == 'resnet50':model = models.resnet50(pretrained=False) elif model_name == 'resnet152':model = models.resnet152(pretrained=False) else:raise Exception("I have not add any models. ")return modelclass CoolSystem(pl.LightningModule):def __init__(self, hparams):super(CoolSystem, self).__init__()self.params = hparams########## define the model ########## model = chose_model(hparams.model)fc_features = model.fc.in_featuresmodel.fc = nn.Linear(fc_features, len(get_label_cate())) self.model = model.to(device)self.loss_fn = nn.CrossEntropyLoss().to(device)self.loss_fn.requires_grad_(True)self.save_hyperparameters()self.resuts = Nonedef forward(self, x):x = self.model(x)return xdef configure_optimizers(self):optimizer = torch.optim.SGD([{'params': self.model.parameters()},], lr=self.params.lr, momentum=self.params.momentum)optimizer = torch.optim.Adam([{'params': self.model.parameters()},], weight_decay=1e-4,lr=self.params.lr)scheduler = torch.optim.lr_scheduler.StepLR(optimizer=optimizer,gamma=0.3, step_size=5)# scheduler = torch.optim.lr_scheduler.CyclicLR(optimizer=optimizer,base_lr=1e-4,max_lr=1e-3,step_size_up=30)scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer=optimizer,T_max=16,eta_min=1e-4)return [optimizer], [scheduler]def training_step(self, batch, batch_idx):"""training"""x, y,img_ori,_ = batchoutput = self.model.forward(x)output = torch.squeeze(output,dim=-1) loss = self.loss_fn(output, y)predicts = F.softmax(output,dim=-1)predicts = torch.max(predicts,dim=-1)[1] acc = (predicts == y).sum().data * 1.0 / self.params.batch_sizeself.log('train_loss', loss, batch_size=batch[0].shape[0], prog_bar=True)self.log('train_acc', acc, batch_size=batch[0].shape[0], prog_bar=True)self.log("lr",self.trainer.optimizers[0].param_groups[0]["lr"], batch_size=batch[0].shape[0], prog_bar=True)# Sample Viz# tumor_index = torch.where(y>0)[0]# for index in tumor_index:# if np.random.randint(1,10)==3:# ran_idx = np.random.randint(1,10)# win = "win_{}".format(ran_idx)# label = y[index]# sample_img = img_ori[index]# title = "label{}_{}".format(label,ran_idx)# visdom_data(sample_img, [], viz=viz_tumor_train,win=win,title=title) # normal_index = torch.where(y==0)[0] # for index in normal_index:# if np.random.randint(1,50)==3:# ran_idx = np.random.randint(1,10)# win = "win_{}".format(ran_idx)# label = y[index]# sample_img = img_ori[index]# title = "label{}_{}".format(label,ran_idx)# visdom_data(sample_img, [], viz=viz_normal_train,win=win,title=title) return {'loss': loss, 'train_acc': acc}def validation_step(self, batch, batch_idx):# OPTIONALx, y,img_ori,_ = batchoutput = self.model.forward(x)output = torch.squeeze(output,dim=-1) loss = self.loss_fn(output, y)predicts = F.softmax(output,dim=-1)predicts = torch.max(predicts,dim=-1)[1] pred_acc_bool = (predicts == y)acc = pred_acc_bool.type(torch.float).sum().data * 1.0 / self.params.batch_size# Calculate the accuracy of each category separatelyall_labes = get_label_cate()results = []for label in all_labes:pred_index = torch.where(predicts==label)[0]acc_cnt = torch.sum(y[pred_index]==label)fail_cnt = torch.sum(y[pred_index]!=label)label_cnt = torch.sum(y==label)results.append([label,acc_cnt.cpu().item(),fail_cnt.cpu().item(),label_cnt.cpu().item()])# Sample Viztumor_index = torch.where(y>0)[0]for index in tumor_index:if np.random.randint(1,10)==3:ran_idx = np.random.randint(1,10)win = "win_{}".format(ran_idx)label = y[index]sample_img = img_ori[index]title = "label{}_{}".format(label,ran_idx)visdom_data(sample_img, [], viz=viz_tumor_valid,win=win,title=title) normal_index = torch.where(y==0)[0] for index in normal_index:if np.random.randint(1,50)==3:ran_idx = np.random.randint(1,10)win = "win_{}".format(ran_idx)label = y[index]sample_img = img_ori[index]title = "label{}_{}".format(label,ran_idx)visdom_data(sample_img, [], viz=viz_normal_valid,win=win,title=title) results = np.array(results)if self.results is None:self.results = results else:self.results = np.concatenate((self.results,results),axis=0)self.log('val_loss', loss, batch_size=batch[0].shape[0], prog_bar=True)self.log('val_acc', acc, batch_size=batch[0].shape[0], prog_bar=True)return {'val_loss': loss, 'val_acc': acc}def on_validation_epoch_start(self):self.results = Nonedef on_validation_epoch_end(self):# For summary calculation# if self.trainer.state.stage==RunningStage.SANITY_CHECKING:# return columns = ["label","acc_cnt","fail_cnt","real_cnt"]results_pd = pd.DataFrame(self.results,columns=columns)all_labes = get_label_cate()for label in all_labes:acc_cnt = results_pd[results_pd["label"]==label]["acc_cnt"].sum()fail_cnt = results_pd[results_pd["label"]==label]["fail_cnt"].sum()real_cnt = results_pd[results_pd["label"]==label]["real_cnt"].sum()self.log('acc_cnt_{}'.format(label), float(acc_cnt), prog_bar=True)self.log('fail_cnt_{}'.format(label), float(fail_cnt), prog_bar=True)self.log('real_cnt_{}'.format(label), float(real_cnt), prog_bar=True)if acc_cnt+fail_cnt==0:acc = 0.0else:acc = acc_cnt/(acc_cnt+fail_cnt)if real_cnt==0:recall = 0.0else:recall = acc_cnt/real_cntself.log('acc_{}'.format(label), acc, prog_bar=True)self.log('recall_{}'.format(label), recall, prog_bar=True)def train_dataloader(self):hparams = self.params# types = hparams.type# split_data_total = []# file_path = hparams.data_path # type# tumor_mask_path = hparams.tumor_mask_path# csv_path = os.path.join(file_path,hparams.train_csv) #type# split_data = pd.read_csv(csv_path).values[:,0].tolist() #type# if split_data_total is None:# split_data_total = split_data# else:# split_data_total = combine(split_data_total,split_data)# wsi_path = os.path.join(file_path,"data") #type# mask_path = os.path.join(file_path,tumor_mask_path) #type# dataset_train = Whole_Slide_Bag_COMBINE(file_path,wsi_path,mask_path,work_type="train",patch_path=hparams.patch_path,# patch_size=hparams.image_size,split_data=split_data,patch_level=hparams.patch_level)dataset_train = Whole_Slide_Bag_COMBINE_togeter(hparams,work_type="train",patch_size=hparams.image_size,patch_level=hparams.patch_level)train_loader = DataLoader(dataset_train,batch_size=self.params.batch_size,collate_fn=self._collate_fn,shuffle=True,num_workers=self.params.num_workers)# data_summarize(train_loader)return train_loaderdef val_dataloader(self):hparams = self.params# types = hparams.type## file_path = hparams.data_path# tumor_mask_path = hparams.tumor_mask_path# csv_path = os.path.join(file_path,hparams.valid_csv)# split_data = pd.read_csv(csv_path).values[:,0].tolist()# wsi_path = os.path.join(file_path,"data")# mask_path = os.path.join(file_path,tumor_mask_path)# dataset_valid = Whole_Slide_Bag_COMBINE(file_path,wsi_path,mask_path,work_type="valid",patch_path=hparams.patch_path,# patch_size=hparams.image_size,split_data=split_data,patch_level=hparams.patch_level,# )dataset_valid = Whole_Slide_Bag_COMBINE_togeter(hparams,work_type='valid',patch_size=hparams.image_size,patch_level=hparams.patch_level)val_loader = DataLoader(dataset_valid,batch_size=self.params.batch_size,collate_fn=self._collate_fn,shuffle=False,num_workers=self.params.num_workers)return val_loaderdef _collate_fn(self,batch):first_sample = batch[0]aggregated = []for i in range(len(first_sample)):if i==0:sample_list = [sample[i] for sample in batch]aggregated.append(torch.stack(sample_list, dim=0))elif i==1:sample_list = [sample[i] for sample in batch]aggregated.append(torch.from_numpy(np.array(sample_list)))else:aggregated.append([sample[i] for sample in batch]) return aggregated def get_last_ck_file(checkpoint_path):list = os.listdir(checkpoint_path)list.sort(key=lambda fn: os.path.getmtime(checkpoint_path+"/"+fn) if not os.path.isdir(checkpoint_path+"/"+fn) else 0) return list[-1]def main(hparams):checkpoint_path = os.path.join(hparams.work_dir,"checkpoints",hparams.model_name)print(checkpoint_path)filename = 'slfcd-{epoch:02d}-{val_loss:.2f}'checkpoint_callback = ModelCheckpoint(monitor='val_loss',dirpath=checkpoint_path,filename=filename,save_top_k=3,auto_insert_metric_name=False) logger_name = "app_log"model_logger = (pl_loggers.TensorBoardLogger(save_dir=hparams.work_dir, name=logger_name, version=hparams.model_name)) log_path = os.path.join(hparams.work_dir,logger_name,hparams.model_name) if hparams.load_weight:file_name = get_last_ck_file(checkpoint_path)checkpoint_path_file = "{}/{}".format(checkpoint_path,file_name)# model = torch.load(checkpoint_path_file) # model = CoolSystem.load_from_checkpoint(checkpoint_path_file).to(device)# trainer = Trainer(resume_from_checkpoint=checkpoint_path_file)trainer = pl.Trainer(max_epochs=hparams.epochs,gpus=1,accelerator='gpu',logger=model_logger,callbacks=[checkpoint_callback],log_every_n_steps=1) trainer.fit(model,ckpt_path=checkpoint_path_file) else:if os.path.exists(checkpoint_path):shutil.rmtree(checkpoint_path)os.mkdir(checkpoint_path)if os.path.exists(log_path):shutil.rmtree(log_path)os.mkdir(log_path)model = CoolSystem(hparams)# data_summarize(model.val_dataloader())trainer = pl.Trainer(max_epochs=hparams.epochs,gpus=1,accelerator='gpu',logger=model_logger,callbacks=[checkpoint_callback],log_every_n_steps=1) trainer.fit(model)def data_summarize(dataloader):it = iter(dataloader)size = len(dataloader)viz_number_tumor = 0viz_number_normal = 0 label_stat = []for index in range(size):img,label,img_ori,item = next(it)img_ori = img_ori.cpu().numpy()[0]type = item['type'][0]if type=="annotation":label_stat.append(item['label'])if viz_number_tumor<10:viz_number_tumor += 1visdom_data(img_ori,[],title="tumor_{}".format(index), viz=viz_tumor_valid)else:label_stat.append(0)if viz_number_normal<10:viz_number_normal += 1 visdom_data(img_ori,[], title="normal_{}".format(index),viz=viz_normal_valid)label_stat = np.array(label_stat)print("label_stat 1:{},2:{},3:{}".format(np.sum(label_stat==1),np.sum(label_stat==2),np.sum(label_stat==3)))if __name__ == '__main__':cnn_path = 'custom/configs/config_together.json'with open(cnn_path, 'r') as f:args = json.load(f) hyperparams = Namespace(**args) main(hyperparams){"model": "resnet50","batch_size": 64,"image_size": 64,"patch_level": 1,"crop_size": 224,"normalize": "True","lr": 0.001,"momentum": 0.9,"cervix":["hsil","lsil"],"data_path": "/home/bavon/datasets/wsi/{cervix}","g_path": "/home/bavon/datasets/wsi","train_csv": "train.csv","valid_csv": "valid.csv","tumor_mask_path": "tumor_mask_level1","patch_path": "patches_level1","data_path_train": "/home/bavon/datasets/wsi/{cervix}/patches/train","data_path_valid": "/home/bavon/datasets/wsi/{cervix}/patches/valid","epochs": 500,"log_every": 5,"num_workers": 5,"load_weight": false,"model_name" : "resnet50_level1","work_dir": "results"
}from __future__ import print_function, division
import os
import torch
import numpy as np
import pandas as pd
import math
import re
import pdb
import pickle
import cv2
import openslide
from torch.utils.data import Dataset, DataLoader, sampler
from torchvision import transforms, utils, models
import torch.nn.functional as Ffrom PIL import Image
import h5py
from random import randrange
from utils.constance import get_tumor_label_catedef eval_transforms(pretrained=False):if pretrained:mean = (0.485, 0.456, 0.406)std = (0.229, 0.224, 0.225)else:mean = (0.5,0.5,0.5)std = (0.5,0.5,0.5)trnsfrms_val = transforms.Compose([transforms.ToTensor(),transforms.Normalize(mean = mean, std = std)])return trnsfrms_valclass Whole_Slide_Bag_COMBINE_togeter(Dataset):"""Custom slide dataset,use multiple wsi file,in which has multiple patches"""def __init__(self,hparams,target_patch_size=-1,custom_downsample=1,patch_level=0,patch_size=256,work_type="train",):"""Args:file_path (string): Path to the .h5 file containing patched data.wsi_path: Path to the .wsi file containing wsi data.mask_path: Path to the mask file containing tumor annotation mask data.custom_downsample (int): Custom defined downscale factor (overruled by target_patch_size)target_patch_size (int): Custom defined image size before embedding"""self.whparams = hparams# self.split_data = []# self.file_path = []self.tumor_mask_path = self.whparams.tumor_mask_pathself.patch_level = patch_levelself.patch_size = patch_sizewsi_data = {}patches_bag_list = []patches_tumor_patch_file_list = []pathces_normal_len = 0pathces_tumor_len = 0file_names = []# self.file_path_global = self.whparams.data_pathself.file_path = self.whparams.g_pathcervixes = self.whparams.cervixfor cervix in cervixes:l_path = self.whparams.data_path.format(cervix = cervix)csv_path = os.path.join(l_path,self.whparams.valid_csv)split_data = pd.read_csv(csv_path).values[:,0].tolist()# wsi_path = os.path.join(self.file_path,"data")mask_path = os.path.join(l_path,self.tumor_mask_path)# loop all patch files,and combine the coords datafor svs_file in split_data:single_name = svs_file.split(".")[0]file_names.append(single_name)patch_file = os.path.join(l_path,self.whparams.patch_path,single_name + ".h5") wsi_file = os.path.join(l_path,"data",svs_file) npy_file = single_name + ".npy"npy_file = os.path.join(mask_path,npy_file) wsi_data[single_name] = openslide.open_slide(wsi_file)scale = wsi_data[single_name].level_downsamples[self.whparams.patch_level]with h5py.File(patch_file, "r") as f:print("patch_file:",patch_file)self.patch_coords = np.array(f['coords'])patch_level = f['coords'].attrs['patch_level']patch_size = f['coords'].attrs['patch_size']# sum data lengthpathces_normal_len += len(f['coords'])if target_patch_size > 0:target_patch_size = (target_patch_size, ) * 2elif custom_downsample > 1:target_patch_size = (patch_size // custom_downsample, ) * 2# Normal patch datafor coord in f['coords']:patches_bag = {"name":single_name,"scale":scale,"type":"normal","cervix":cervix} patches_bag["coord"] = np.array(coord) /scalepatches_bag["coord"] = patches_bag["coord"].astype(np.int16)patches_bag["patch_level"] = patch_levelpatches_bag["label"] = 0patches_bag_list.append(patches_bag)# Annotation patch datafor label in get_tumor_label_cate():if work_type=="train":# Using augmentation image for validationpatch_img_path = os.path.join(l_path,"tumor_patch_img",str(label),"origin")else:# Using origin image for validationpatch_img_path = os.path.join(l_path,"tumor_patch_img",str(label),"origin")file_list = os.listdir(patch_img_path)for file in file_list:if not single_name in file:continuetumor_file_path = os.path.join(patch_img_path,file)patches_tumor_patch_file_list.append(tumor_file_path)pathces_tumor_len += 1self.patches_bag_list = patches_bag_listself.pathces_normal_len = pathces_normal_len self.patches_tumor_patch_file_list = patches_tumor_patch_file_listself.pathces_tumor_len = pathces_tumor_lenself.pathces_total_len = pathces_tumor_len + pathces_normal_lenself.roi_transforms = eval_transforms()self.target_patch_size = target_patch_sizedef __len__(self):return self.pathces_total_lendef __getitem__(self, idx):# Judge type by index valueif idx>=self.pathces_normal_len:# print("mask_tumor_size is:{},coord:{}".format(mask_tumor_size,coord))file_path = self.patches_tumor_patch_file_list[idx-self.pathces_normal_len]t = file_path.split("/")try:label = int(t[-3])# label = 1except Exception as e:print("sp err:{}".format(t))img_ori = cv2.imread(file_path)item = {}else:item = self.patches_bag_list[idx]name = item['name']scale = item['scale']coord = item['coord']cervix = item['cervix']wsi_file = os.path.join(self.file_path,cervix,"data",name + ".svs") wsi = openslide.open_slide(wsi_file) # read image from wsi with coordination coord_ori = (coord * scale).astype(np.int16) img_ori = wsi.read_region(coord_ori, self.patch_level, (self.patch_size, self.patch_size)).convert('RGB')img_ori = cv2.cvtColor(np.array(img_ori), cv2.COLOR_RGB2BGR) label = 0if self.target_patch_size > 0 :img_ori = img_ori.resize(self.target_patch_size)img = self.roi_transforms(img_ori)return img,label,img_ori,item