写在前面
同上一篇的PCoA分析,这个也是基于公司结果基础上的再次分析,重新挑选样本,在公司结果提供的csv结果表上进行删减,本地重新分析作图
步骤
表格预处理
- 在公司给的ClusterTree的原始表格数据里选取要保留的样本,同样保存为逗号分隔的csv文件
代码演示
无色版
install.packages('vegan')
install.packages('dendextend')
install.packages('circlize')data <- read.table('F:\\Analysis\\RA_Sanhe cow\\Microgenome\\Cluster_Tree\\table.g10.cluster.csv', header=T, sep = ',', check.names=FALSE)
rownames(data)=data[,1]
data=data[,-1]
library(vegan)
data=decostand(data, MARGIN=2, "total")
otu=t(data)
otu_dist=vegdist(otu, method="bray", diag=TRUE, upper=TRUE, p=2)
#进行聚类分析并作图
hclust=hclust(otu_dist, method="average")
plot(hclust)library(dendextend)
library(circlize)
tree=as.dendrogram(hclust)
par(mfrow=c(2,2), mar=c(3,3,1,5), cex=0.7)
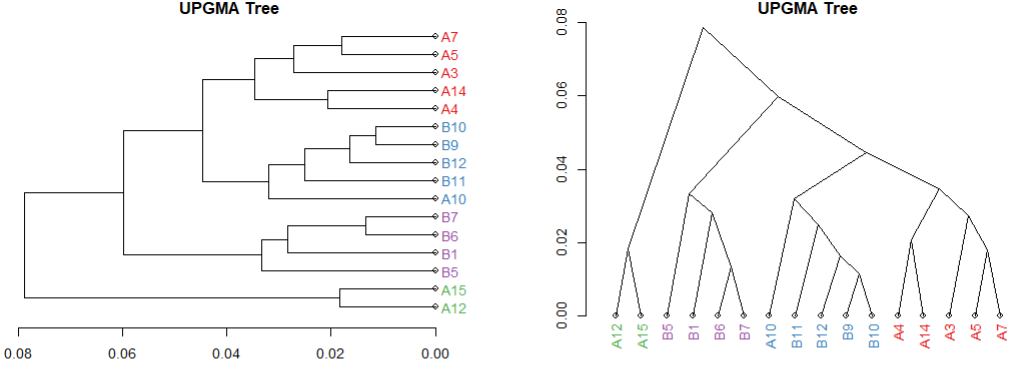
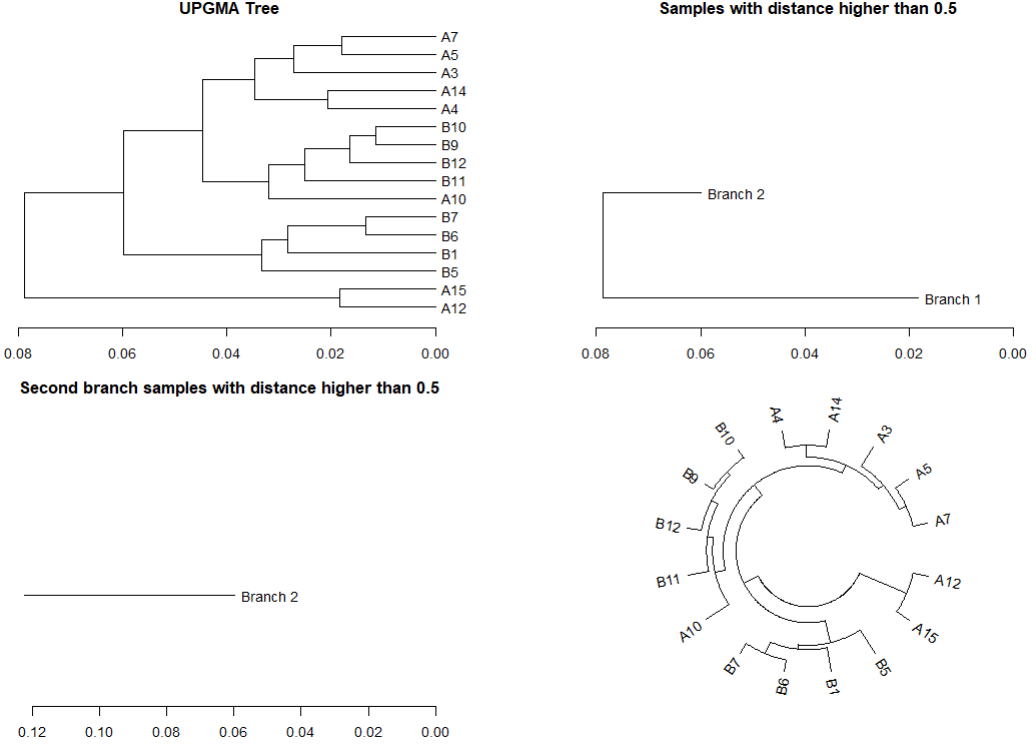
plot(tree, horiz=TRUE, main="UPGMA Tree")
#隐藏平均距离小于0.5的对象(类群)
plot(cut(tree, h=0.5)$upper, horiz=TRUE, main="Samples with distance higher than 0.5")
#在上一步基础上筛选第二个分类簇
plot(cut(tree, h=0.5)$upper[[2]], horiz=TRUE, main="Second branch samples with distance higher than 0.5")
circlize_dendrogram(tree)
上色版
##完整代码
data <- read.table('F:\\Analysis\\RA_Sanhe cow\\Microgenome\\Cluster_Tree\\Demo_ClusterTree.csv', header=T, sep = ',', check.names=FALSE)
rownames(data)=data[,1]
data=data[,-1]
library(vegan)
data=decostand(data, MARGIN=2, "total")
otu=t(data)
otu_dist=vegdist(otu, method="bray", diag=TRUE, upper=TRUE, p=2)
#进行聚类分析并作图
hclust=hclust(otu_dist, method="average")
library(dendextend)
library(RColorBrewer)
hcd=as.dendrogram(hclust)
labelColors=brewer.pal(n=4, name="Set1")
#聚类分组,预设聚类簇数目为4
clusMember=cutree(hcd, 4)
#自定义函数,根据聚类结果进行着色,4 种颜色
colLab=function(n) {if (is.leaf(n)) {a=attributes(n)labCol=labelColors[clusMember[which(names(clusMember)==a$label)]]attr(n, "nodePar")=c(a$nodePar, lab.col=labCol)}n
}
clusDendro=dendrapply(hcd, colLab)
plot(clusDendro, main ="UPGMA Tree", type="rectangle", horiz=TRUE)
plot(clusDendro, main ="UPGMA Tree", type="triangle")