介绍
科研绘图系列:python语言绘制SCI图合集
加载python
import numpy as np
import pandas as pd import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as snsfrom statsmodels.stats.multitest import multipletests# Setup for local running - please delete this block
import sys
sys.path.append('C:\\Users\\ncaptier\\Documents\\GitHub\\multipit\\')from multipit.result_analysis.plot import plot_metrics
from utils import plot_average_perf, plot_benchmark, change_width, annotate, reshape_clustermap
数据下载
python语言绘图合集:
- 百度网盘链接: 从百度网盘下载数据
- 提取码: 科研绘图系列:python语言绘制SCI图合集

代码
加载python
import numpy as np
import pandas as pd import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import seaborn as snsfrom statsmodels.stats.multitest import multipletests# Setup for local running - please delete this block
import sys
sys.path.append('C:\\Users\\ncaptier\\Documents\\GitHub\\multipit\\')from multipit.result_analysis.plot import plot_metrics
from utils import plot_average_perf, plot_benchmark, change_width, annotate, reshape_clustermap
Figures 2, s7 - s10
rna_os = {"XGboost": 0.663, "LR": 0.627, "RF": 0.624, "Cox": 0.542}
rna_pfs = {"XGboost": 0.637, "LR": 0.663, "RF": 0.591, "Cox": 0.569}clinical_os = {"XGboost": 0.579, "LR": 0.667, "RF": 0.652, "Cox": 0.631}
clinical_pfs = {"XGboost": 0.552, "LR": 0.575, "RF": 0.563, "Cox": 0.526}radiomics_os = {"XGboost": 0.574, "LR": None, "RF": 0.556, "Cox": 0.563}
radiomics_pfs = {"XGboost": 0.634, "LR": 0.568, "RF": 0.565, "Cox": 0.58}pathomics_os = {"XGboost": 0.547, "LR": 0.54, "RF": 0.561, "Cox": 0.538}
pathomics_pfs = {"XGboost": 0.588, "LR": 0.573, "RF": 0.534, "Cox": 0.524}shap_rna_xgboost = pd.read_csv("..\\source_data\\unimodal_shapvalues\\1year_death\\XGboost_RNA.csv", index_col=0)
shap_rna_LR = pd.read_csv("..\\source_data\\unimodal_shapvalues\\1year_death\\LR_RNA.csv", index_col=0)
shap_rna_Cox = pd.read_csv("..\\source_data\\unimodal_shapvalues\\OS\\Cox_RNA.csv", index_col=0)
shap_rna_RF = pd.read_csv("..\\source_data\\unimodal_shapvalues\\OS\\RF_RNA.csv", index_col=0)correlation_signs = pd.read_csv("..\\source_data\\unimodal_shapvalues\\shap_features_correlations\\RNA_signs_os.csv", index_col=0)
correlation_signs.head()
consensus_features = (correlation_signs.sum(axis=1) == -4) | (correlation_signs.sum(axis=1) == 4)# Note: For radiomics OS, replace -4 and 4 by -3 and 3 (only 3 algorithms out of 4 taken into account)df_rank = pd.concat([np.abs((shap_rna_xgboost.iloc[:, :-1].T)[consensus_features]).mean(axis=1).rank(ascending=True).rename("XGBoost"),np.abs((shap_rna_LR.iloc[:, :-1].T)[consensus_features]).mean(axis=1).rank(ascending=True).rename("LR"),np.abs((shap_rna_RF.iloc[:, :-1].T)[consensus_features]).mean(axis=1).rank(ascending=True).rename("RF"),np.abs((shap_rna_Cox.iloc[:, :-1].T)[consensus_features]).mean(axis=1).rank(ascending=True).rename("Cox")],axis=1
)df_rank.head()
weights = [val - 0.5 for val in rna_os.values()]final_importance = (df_rank.apply(lambda row: (1/sum(weights))*(weights[0]*row["XGBoost"] + weights[1]*row["LR"] + weights[2]*row["RF"] + weights[3]*row["Cox"]), axis=1).sort_values(ascending=False))final_importance = (final_importance/df_rank.shape[0]) * (correlation_signs[consensus_features]["XGboost"].loc[final_importance.index])
final_importance = final_importance.to_frame().rename(columns={0: "Consensus importance"})
final_importance["Impact"] = 1*(final_importance["Consensus importance"] > 0)
final_importance = final_importance.replace(to_replace = {"Impact": {0: "Lower risk", 1: "Increase risk"}})final_importance.head(5)
fig, ax = plt.subplots(figsize=(10, 8))sns.barplot(data=final_importance.reset_index(),orient="h",x="Consensus importance",y="index",hue="Impact",palette=["blue", "red"],dodge=False,ax=ax)ax.set(xlabel=None, ylabel=None)
ax.set_axisbelow(True)
ax.yaxis.grid(color="gray", linestyle="dashed")
ax.legend(fontsize=12)
ax.axvline(x=0, color="k")
ax.xaxis.set_tick_params(labelsize=12)
ax.yaxis.set_tick_params(labelsize=12)
ax.set_title("Consensus importance", fontsize=14)
ax.set_xlim(-1.05, 1.05)
plt.tight_layout()
sns.despine()

Figures 3, s14, s16 - s23
results_cl = pd.read_csv("..\\source_data\\multimodal_performance\\1year_death\\late_XGboost.csv", index_col=0)
results_cl.head(5)
fig = plot_metrics(results_cl,metrics="roc_auc",models = list(results_cl.columns[1:]),annotations = {"1 modality": (0, 3), "2 modalities": (4, 9), "3 modalities": (10, 13), "4 modalities": (14, 15)},title=None,ylim=(0.5, 0.86),y_text=0.85,ax=None)
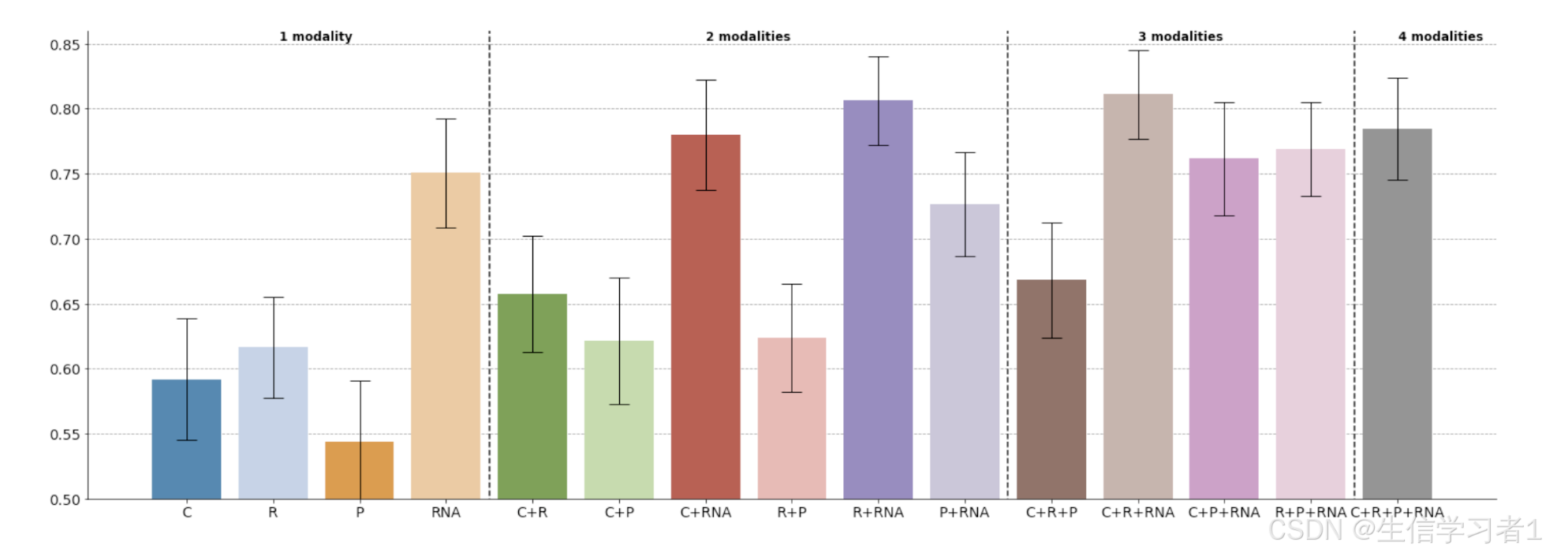
results_surv = pd.read_csv("..\\source_data\\multimodal_performance\\OS\\late_RF.csv", index_col=0)
results_surv.head(5)fig = plot_metrics(results_surv,metrics='c_index',models = list(results_surv.columns[1:]),annotations = {"1 modality": (0, 3), "2 modalities": (4, 9), "3 modalities": (10, 13), "4 modalities": (14, 15)},title=None, ylim=(0.5, 0.77), y_text=0.77,ax=None)
Figure 4
marginal_contributions = pd.read_csv("..\\source_data\\marginal_contributions_latefus.csv", index_col=0)
marginal_contributions.head()
cmap_TN = sns.clustermap(marginal_contributions[marginal_contributions["results"]=="TN"][["C", "R", "RNA"]],cmap="bwr",center=0,yticklabels=False,xticklabels=False,vmin=-0.08,vmax=0.14)
reshape_clustermap(cmap_TN, cell_width=0.25, cell_height=0.015)
px = 1/plt.rcParams['figure.dpi'] fig, ax = plt.subplots(figsize=(100*px, 675*px))
ax.set_xlim(-0.15, 0)(marginal_contributions.loc[cmap_TN.data2d.index, "multi_pred"].iloc[::-1] - 0.5).plot.barh(width=0.85, ax=ax, color="k")sns.despine()
Figure 5
models = ['late_XGboost', 'late_LR', 'early_XGboost', 'early_select_XGboost','early_LR', 'early_select_LR', 'dyam', 'dyam_optim', 'dyam_select', 'dyam_optim_select']algorithms = ["XGboost", "LR", "XGboost", "XGboost", "LR", "LR", "Perceptron", "Perceptron", "Perceptron", "Perceptron"]#To save best multimodal models
list_best_1y , list_names_best_1y = [], []
list_best_6m, list_names_best_6m = [], []#To save unimodal models
list_clinical_1y, list_clinical_6m = [], []
list_radiomics_1y, list_radiomics_6m = [], []
list_pathomics_1y, list_pathomics_6m = [], []
list_RNA_1y, list_RNA_6m = [], []for mod in models:# Collect unimodal models and best multimodal models for 1-year death predictionresults_1y = pd.read_csv("..\\source_data\\multimodal_performance\\1year_death\\" + mod + ".csv", index_col=0)list_clinical_1y.append(results_1y["C"])list_radiomics_1y.append(results_1y["R"])list_pathomics_1y.append(results_1y["P"])list_RNA_1y.append(results_1y["RNA"])best_1y = results_1y.iloc[:, 1:].mean().idxmax()list_best_1y.append(results_1y[best_1y].rename(mod))list_names_best_1y.append(best_1y)# Collect unimodal models and best multimodal models for 6-months progression predictionresults_6m = pd.read_csv("..\\source_data\\multimodal_performance\\6months_progression\\" + mod + ".csv", index_col=0)list_clinical_6m.append(results_6m["C"])list_radiomics_6m.append(results_6m["R"])list_pathomics_6m.append(results_6m["P"])list_RNA_6m.append(results_6m["RNA"])best_6m = results_6m.iloc[:, 1:].mean().idxmax()list_best_6m.append(results_6m[best_6m].rename(mod))list_names_best_6m.append(best_6m)# Concatenate best multimodal models accross integration strategies
results_multimodal_1y = pd.concat(list_best_1y, axis=1)
results_multimodal_1y["metric"] = "1y death AUC"results_multimodal_6m = pd.concat(list_best_6m, axis=1)
results_multimodal_6m["metric"] = "6m progression AUC"results_multimodal = pd.concat([results_multimodal_1y, results_multimodal_6m], axis=0)# Select best unimodal models accross predictive algorithms
best_clinical_1y = np.argmax([mod.mean() for mod in list_clinical_1y])
best_radiomics_1y = np.argmax([mod.mean() for mod in list_radiomics_1y])
best_pathomics_1y = np.argmax([mod.mean() for mod in list_pathomics_1y])
best_RNA_1y = np.argmax([mod.mean() for mod in list_RNA_1y])
results_unimodal_1y = pd.concat([list_clinical_1y[best_clinical_1y],list_radiomics_1y[best_radiomics_1y],list_pathomics_1y[best_pathomics_1y],list_RNA_1y[best_RNA_1y]],axis=1)
results_unimodal_1y["metric"] = "1y death AUC"best_clinical_6m = np.argmax([mod.mean() for mod in list_clinical_6m])
best_radiomics_6m = np.argmax([mod.mean() for mod in list_radiomics_6m])
best_pathomics_6m = np.argmax([mod.mean() for mod in list_pathomics_6m])
best_RNA_6m = np.argmax([mod.mean() for mod in list_RNA_1y])
results_unimodal_6m = pd.concat([list_clinical_6m[best_clinical_6m],list_radiomics_6m[best_radiomics_6m],list_pathomics_6m[best_pathomics_6m],list_RNA_6m[best_RNA_6m]],axis=1)
results_unimodal_6m["metric"] = "6m progression AUC"results_unimodal = pd.concat([results_unimodal_1y, results_unimodal_6m], axis=0)_, ax = plt.subplots(figsize=(25, 10))annotations = {0: algorithms[best_clinical_1y], 1: algorithms[best_clinical_6m],2: algorithms[best_radiomics_1y], 3: algorithms[best_radiomics_6m],4: algorithms[best_pathomics_1y], 5: algorithms[best_pathomics_6m],6: algorithms[best_RNA_1y], 7: algorithms[best_RNA_6m]}fig = plot_benchmark(results_unimodal, metrics = ["1y death AUC", "6m progression AUC"], new_width = 0.15,annotations = annotations ,ylim = (0.5, 0.86), title = "Best unimodal models",ax=ax)plt.tight_layout()
_, ax = plt.subplots(figsize=(25, 10))annotations = {i: list_names_best_1y[i//2].split('+') if i%2 == 0 else list_names_best_6m[i//2].split('+') for i in range(20)}fig = plot_benchmark(results_multimodal, metrics = ["1y death AUC", "6m progression AUC"], new_width = 0.07,annotations = annotations ,ylim = (0.5, 0.86), title = "Best multimodal combination for different integration strategies",ax=ax)plt.tight_layout()
Figure 6, s15
models = ['late_XGboost', 'late_LR', 'early_XGboost', 'early_select_XGboost', 'early_LR', 'early_select_LR', 'dyam', 'dyam_select']list_average = []
list_std = []for mod in models:results = pd.read_csv("..\\source_data\\multimodal_performance\\1year_death\\" + mod + ".csv", index_col=0)results_agg = pd.DataFrame(index = results.index)results_agg["1 modality"] = results[["C", "R", "P", "RNA"]].mean(axis=1)results_agg["2 modalities"] = results[["C+R", "C+P", "C+RNA", "R+P", "P+RNA", "R+RNA"]].mean(axis=1)results_agg["3 modalities"] = results[["C+R+P", "C+R+RNA", "C+P+RNA", "R+P+RNA"]].mean(axis=1)results_agg["4 modalities"] = results["C+R+P+RNA"].copy()list_average.append(results_agg.mean().rename(mod)) list_std.append(results_agg.std().rename(mod))data = pd.concat(list_average, axis=1).T
data_std = pd.concat(list_std, axis=1).T.values.flatten(order="F")data
fig = plot_average_perf(data,data_std,markers = ["o", "*", "^", "s", "X","8", "D", "h", "P", ">"],sizes = [10, 16, 12, 10, 10, 10, 10, 10, 10, 12],ylim = (0.5, 0.81))plt.tight_layout()
Figure 7
list_sig = ['CRMA', 'CTLA4', 'CX3CL1', 'CXCL9', 'CYT', 'EIGS', 'ESCS', 'FTBRS', 'HLADRA', 'HRH1', 'IFNgamma', 'IMPRES', 'IRG', 'Immunopheno', 'MPS', 'PD1', 'PDL1','PDL2', 'Renal101', 'TIG', 'TLS', 'TME']list_gsea = ['APM', 'CECMup', 'CECMdown', 'IIS', 'IMS', 'IPRES', 'MFP', 'MIAS', 'PASSPRE', 'TIS']list_deconv = ['CD8T_CIBERSORT', 'CD8T_MCPcounter', 'CD8T_Xcell', 'Immuno_CIBERSORT']
results_1yeardeath = pd.read_csv("..\\source_data\\signatureRNA_performance\\1year_death\\signatures_RNA.csv", index_col=0)temp = pd.read_csv("..\\source_data\\multimodal_performance\\1year_death\\late_XGboost.csv", index_col=0)results_1yeardeath["best_RNA"] = temp["RNA"]
results_1yeardeath["best_multimodal"] = temp["C+R+RNA"]results_1yeardeath.head(5)
list_sig_sorted = list(results_1yeardeath[list_sig].mean().apply(lambda x: max(x, 1-x)).sort_values().index)
list_gsea_sorted = list(results_1yeardeath[list_gsea].mean().apply(lambda x: max(x, 1-x)).sort_values().index)
list_deconv_sorted = list(results_1yeardeath[list_deconv].mean().apply(lambda x: max(x, 1-x)).sort_values().index)results_1yeardeath = results_1yeardeath[["metric"] + list_sig_sorted + list_gsea_sorted + list_deconv_sorted + ["best_RNA", "best_multimodal"]]
color_dic = {}
for col in results_1yeardeath.columns[1:-2]:if results_1yeardeath[col].mean() < 0.5:results_1yeardeath[col] = 1 - results_1yeardeath[col]color_dic[col] = "blue"else:color_dic[col] = "red"color_dic["best_RNA"] = "red"
color_dic["best_multimodal"] = "red"
_, ax = plt.subplots(figsize=(20, 10))fig = plot_metrics(results_1yeardeath,metrics="roc_auc",models = list(results_1yeardeath.columns[1:]),colors = color_dic,ylim=(0.5, 0.86),annotations={"Marker genes \n methods": (0, 21), "GSEA \n methods": (22, 31), "Deconvolution \n methods": (32, 35), "Our ML\n methods": (37,39)},y_text = 0.82,ax = ax)t = plt.setp(ax.get_xticklabels(), rotation=45, ha="right", rotation_mode="anchor")red_patch = mpatches.Patch(color='red', label='Poor prognosis')
blue_patch = mpatches.Patch(color='blue', label='Good prognosis')
ax.legend(handles=[red_patch, blue_patch], fontsize=16)
plt.tight_layout()
Figures 8, s24
pval_OS = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\OS\\pvalues.csv", index_col=0).T
pval_OS["Predictive task"] = "OS"pval_PFS = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\PFS\\pvalues.csv", index_col=0).T
pval_PFS["Predictive task"] = "PFS"pval_1y = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\1year_death\\pvalues.csv", index_col=0).T
pval_1y["Predictive task"] = "1y-death"pval_1y_thr = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\1year_death\\pvalues_threshold.csv", index_col=0).T
pval_1y_thr["Predictive task"] = "1y-death (threshold)"pval_6m = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\6months_progression\\pvalues.csv", index_col=0).T
pval_6m["Predictive task"] = "6m-progression"pval_6m_thr = pd.read_csv("..\\source_data\\multimodal_risk_stratification\\6months_progression\\pvalues_threshold.csv", index_col=0).T
pval_6m_thr["Predictive task"] = "6m-progression (threshold)"
d = {col: "multimodal" for col in pval_OS.columns if len(col.split('+')) > 1}
d.update({"C": "clinical", "R": "Radiomic", "P": "Pathomic",})final_1 = (pd.concat([pval_OS, pval_PFS], axis=0).melt(id_vars = ["Predictive task"]).replace({"variable": d}))
final_2 = (pd.concat([pval_1y, pval_1y_thr, pval_6m, pval_6m_thr], axis=0).melt(id_vars = ["Predictive task"]).replace({"variable":d}))results = pd.concat([final_1, final_2]).reset_index(drop=True)corrected = multipletests(list(results["value"].fillna(1).values), alpha=0.05, method='fdr_bh')results["value"] = -np.log10(corrected[1])results.head()
fig, ax = plt.subplots(figsize=(10, 8))data = results[results["variable"] == "multimodal"]sns.boxplot(data = data, x="variable", y="value", hue="Predictive task", ax=ax)
sns.stripplot(data = data, x="variable", y="value", hue="Predictive task", ax=ax, dodge=True, palette='dark:k', size=4)
sns.despine()handles, labels = ax.get_legend_handles_labels()
ax.legend(handles[:6], labels[:6], fontsize=16, bbox_to_anchor = [0.62, 0.785])ax.set_ylim(-0.1, 7)
ax.set(xlabel=None)
ax.set_ylabel("-log10 pvalue", fontsize=16)
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed')
ax.tick_params(axis='y', labelsize=16)
ax.tick_params(axis='x', labelsize=16)
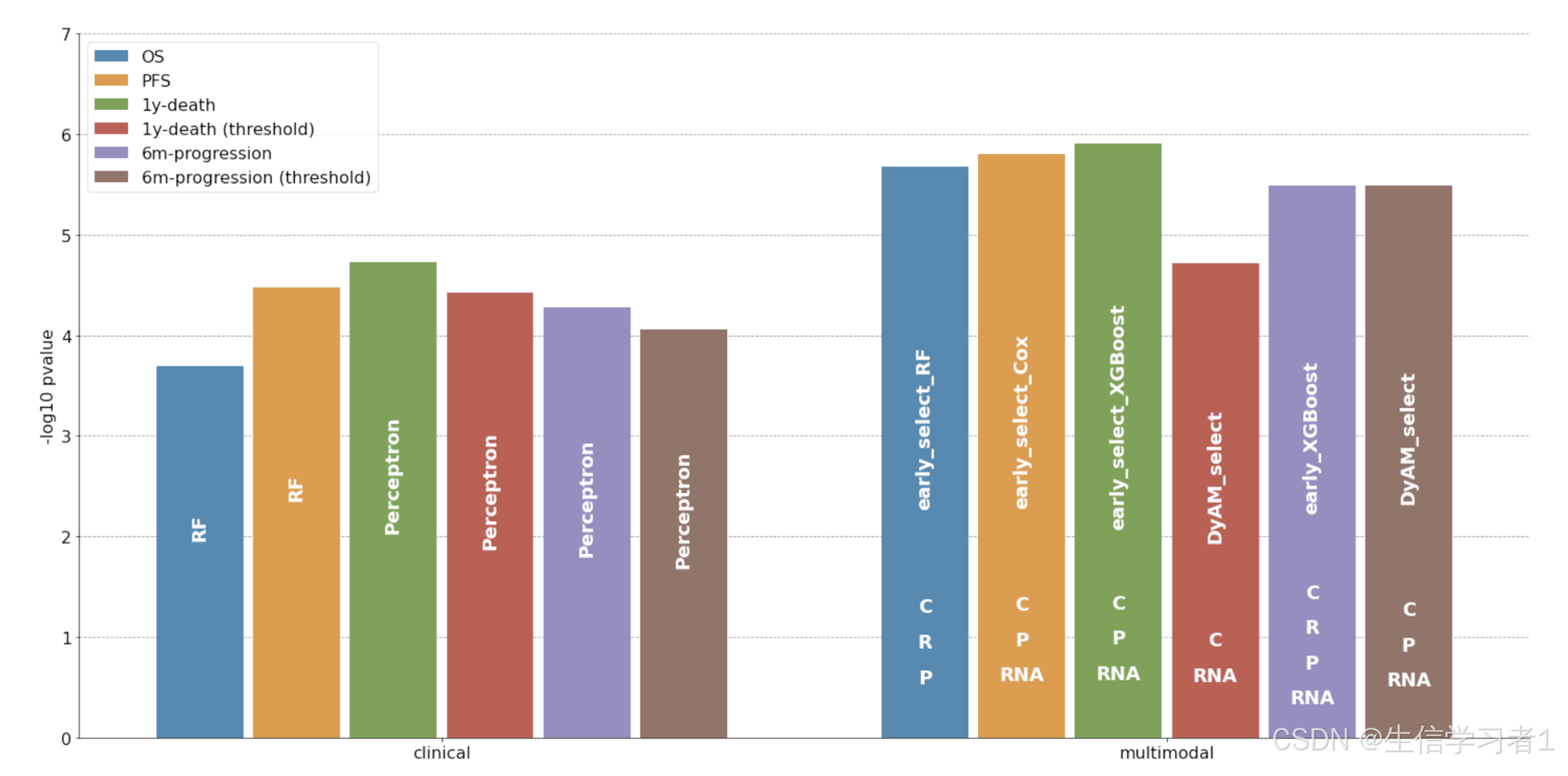
fig, ax = plt.subplots(figsize=(20, 10))
sns.barplot(data = results, x="variable", y="value", hue="Predictive task", ax=ax, estimator=max, err_kws={'linewidth': 0})
sns.despine()
change_width(ax, 0.12)annotations = {0: "RF", 1: "early_select_RF",2: "RF", 3: "early_select_Cox",4: "Perceptron", 5: "early_select_XGBoost",6: "Perceptron", 7: "DyAM_select",8: "Perceptron", 9: "early_XGBoost",10: "Perceptron", 11: "DyAM_select"}annotations_bis = {0: "", 1: ["C", "R", "P"],2: "", 3: ["C", "P", "RNA"],4: "", 5: ["C", "P", "RNA"],6: "", 7: ["C", "RNA"],8: "", 9: ["C", "R", "P", "RNA"],10: "", 11: ["C", "P", "RNA"]}annotate(ax, annotations, rotation = "vertical")
annotate(ax, annotations_bis, position = lambda x: x/6, gap=0.35)ax.legend(fontsize=16)ax.set_ylim(0, 7)
ax.set(xlabel=None)
ax.set_ylabel("-log10 pvalue", fontsize=16)
ax.set_axisbelow(True)
ax.yaxis.grid(color='gray', linestyle='dashed')
ax.tick_params(axis='y', labelsize=16)
ax.tick_params(axis='x', labelsize=16)plt.tight_layout()