实验介绍
分类和回归之间的主要区别在于,在分类中,我们的预测目标是离散的类别,而在回归中,预测目标是连续的预测值。
本实验的研究区域位于佛蒙特州的埃塞克斯郡,使用训练数据来模拟土壤氧化还原深度,然后生成准确度评估图表和统计数据。(数据仅供实验使用,不代表真实值)
实验目标
-
随机森林回归
-
GEE 图表绘制
实验数据
VT_boundary.shp – shapefile 表示感兴趣的示例区域
VT_pedons.shp – 佛蒙特州埃塞克斯县训练数据的 shapefile
var VT_boundary = ee.FeatureCollection("projects/ee-yelu/assets/VT_boundary"),VT_pedons = ee.FeatureCollection("projects/ee-yelu/assets/essex_pedons_all");
实验环境
Chrome浏览器
earth engine账号
目录
第 1 部分:合成时间序列多参数影像数据
第 2 部分:准备训练/验证数据
第 3 部分:运行随机森林回归
第 4 部分:向地图添加回归,创建图例
第 5 部分:创建模型评估统计数据和图表
第 6 部分:验证
第 7 部分:导出
第 8 部分:讨论
时间序列Sentinel-1、Sentinel-2影像预处理
上传矢量数据到earth engine
确保您已将VT_boundary.shp文件上传到您的assets文件夹并将其导入到您的脚本中。确保将其从table重命名为VT_boundary,然后将代码保存到本地存储库。
多时相Sentinel-2影像预处理
因为研究区域位于不同的地理区域,因此使用earth engine 加载自定义矢量时
需要准确地定义矢量文件的投影。
// 研究区
var roi = VT_boundary.union();
Map.addLayer(roi, {}, 'shp', false);
var crs = 'EPSG:4326'; // EPSG number for output projection. 32618 = WGS84/UTM Zone 18N. For more info- http://spatialreference.org/ref/epsg/
S2 影像去云与合成
function maskS2clouds(image) {var qa = image.select('QA60');// Bits 10 and 11 are clouds and cirrus, respectively.var cloudBitMask = 1 << 10;var cirrusBitMask = 1 << 11;// Both flags should be set to zero, indicating clear conditions.var mask = qa.bitwiseAnd(cloudBitMask).eq(0).and(qa.bitwiseAnd(cirrusBitMask).eq(0));return image.updateMask(mask).divide(10000);
}
计算多种植被指数作为特征
// NDVI
function NDVI(img) {var ndvi = img.expression("(NIR-R)/(NIR+R)", {"R": img.select("B4"),"NIR": img.select("B8"),});return img.addBands(ndvi.rename("NDVI"));
}// NDWI
function NDWI(img) {var ndwi = img.expression("(G-MIR)/(G+MIR)", {"G": img.select("B3"),"MIR": img.select("B8"),});return img.addBands(ndwi.rename("NDWI"));
}// NDBI
function NDBI(img) {var ndbi = img.expression("(SWIR-NIR)/(SWIR-NIR)", {"NIR": img.select("B8"),"SWIR": img.select("B12"),});return img.addBands(ndbi.rename("NDBI"));
}//SAVI
function SAVI(image) {var savi = image.expression('(NIR - RED) * (1 + 0.5)/(NIR + RED + 0.5)', {'NIR': image.select('B8'),'RED': image.select('B4')}).float();return image.addBands(savi.rename('SAVI'));
}//IBI
function IBI(image) {// Add Index-Based Built-Up Index (IBI)var ibiA = image.expression('2 * SWIR1 / (SWIR1 + NIR)', {'SWIR1': image.select('B6'),'NIR': image.select('B5')}).rename(['IBI_A']);var ibiB = image.expression('(NIR / (NIR + RED)) + (GREEN / (GREEN + SWIR1))', {'NIR': image.select('B8'),'RED': image.select('B4'),'GREEN': image.select('B3'),'SWIR1': image.select('B11')}).rename(['IBI_B']);var ibiAB = ibiA.addBands(ibiB);var ibi = ibiAB.normalizedDifference(['IBI_A', 'IBI_B']);return image.addBands(ibi.rename(['IBI']));
}//RVI
function RVI(image) {var rvi = image.expression('NIR/Red', {'NIR': image.select('B8'),'Red': image.select('B4')});return image.addBands(rvi.rename('RVI'));
}//DVI
function DVI(image) {var dvi = image.expression('NIR - Red', {'NIR': image.select('B8'),'Red': image.select('B4')}).float();return image.addBands(dvi.rename('DVI'));
}
逐月合成Sentinel-2、Sentinel-1影像,并计算植被指数(通过for循环实现逐月合成,可以根据需要修改为自定义的时间序列)
// 创建var inStack = ee.Image()
for (var i = 0; i < 3; i += 1) {var start = ee.Date('2019-03-01').advance(30 * i, 'day');print(start)var end = start.advance(30, 'day');var dataset = ee.ImageCollection('COPERNICUS/S2_SR').filterDate(start, end).filterBounds(roi).filter(ee.Filter.lt('CLOUDY_PIXEL_PERCENTAGE',75)).map(maskS2clouds).map(NDVI).map(NDWI).map(NDBI).map(SAVI).map(IBI).map(RVI).map(DVI);var inStack_monthly = dataset.median().clip(roi);// 可视化var visualization = {min: 0.0,max: 0.3,bands: ['B4', 'B3', 'B2'],};Map.addLayer(inStack_monthly, visualization, 'S2_' + i, false);// // 纹理特征var B8 = inStack_monthly.select('B8').multiply(100).toInt16();var glcm = B8.glcmTexture({size: 3});var contrast = glcm.select('B8_contrast');var var_ = glcm.select('B8_var');var savg = glcm.select('B8_savg');var dvar = glcm.select('B8_dvar');inStack_monthly = inStack_monthly.addBands([contrast, var_, savg, dvar])// Sentinel-1 var imgVV = ee.ImageCollection('COPERNICUS/S1_GRD').filter(ee.Filter.listContains('transmitterReceiverPolarisation', 'VV')).filter(ee.Filter.listContains('transmitterReceiverPolarisation', 'VH')).filter(ee.Filter.eq('instrumentMode', 'IW')).filterBounds(roi).map(function(image) {var edge = image.lt(-30.0);var maskedImage = image.mask().and(edge.not());return image.updateMask(maskedImage);});var img_S1_asc = imgVV.filter(ee.Filter.eq('orbitProperties_pass', 'ASCENDING'));var VV_img = img_S1_asc.filterDate(start, end).select("VV").median().clip(roi);var VH_img = img_S1_asc.filterDate(start, end).select("VH").median().clip(roi);inStack_monthly = inStack_monthly.addBands([VV_img, VH_img])print("img_S2_monthly", inStack_monthly)inStack = inStack.addBands(inStack_monthly.select(inStack_monthly.bandNames()))
}inStack = inStack.select(inStack.bandNames().remove("constant"))
print(inStack)
在控制台上输出,堆叠后的多波段image:
print("Predictor Layers:", inStack);
inStack.reproject(crs, null, 30);
Map.centerObject(inStack)
单击运行并耐心等待(数据量比较大因此耗时较长)一个名为“ Predictor Layers ”的image对象将出现在控制台中。单击“ Image ”旁边的向下箭头,然后单击“ bands ”旁边的向下箭头以检查此对象。可以看出,我们创建了多时相多参数的遥感影像
准备训练/验证数据
A. 加载 AOI pedons shapefile
在开始之前,需要将样本数据VT_pedons.shp作为assets加载到 GEE ,并导入到我们的代码中,以便接下来在回归中使用。
转到左侧面板中的assets选项卡,找到shapefile,然后单击它,此时会弹出预览:

导航到“features”选项卡并浏览 shapefile 的不同属性。本实验目标预测土壤氧化还原深度,因此“ REDOX_CM ”是我们需要预测的变量。
单击右下角的蓝色IMPORT按钮将其添加到我们的代码中。
此时会看到一个table变量已添加到顶部的导入列表中。我们将新 shapefile 的名称从默认的table更改为VT_pedons。
现在您应该有两个import:VT_boundary和VT_pedons。
B. 使用 pedons 准备训练和验证数据
“VT_pedons”是用于回归的样本数据,我们现在将其转为ee.FeatureCollection()类型,以便我们在GEE中调用。
var trainingFeatureCollection = ee.FeatureCollection(VT_pedons, 'geometry');
接下来我们开始用随机森林做回归
运行随机森林回归
A. 选择需要使用的波段
出于本练习的目的,我们刚刚堆叠了多个波段,然而在一些研究中,可能仅需要某些波段参与回归,因此可以通过
ee.Image().select("波段名称")
来筛选需要的波段。
这里,我们仍然使用全部的波段进行回归分析。
var bands = inStack.bandNames(); //All bands on included here
B.提取训练数据
我们的下一步是根据样本点坐标,提取影像上相应位置的多个波段的数据。
var training = inStack.reduceRegions({collection:trainingFeatureCollection,reducer :ee.Reducer.mean(),scale:100,tileScale :5,crs: crs
});
print(training)
var trainging2 = training.filter(ee.Filter.notNull(bands));
注意:我们将训练数据的空间分辨率(scale)设置为 30 m——使其与我们之前应用的预测层重投影保持一致。
如果有兴趣进一步探索sampleRegions命令,只需在左侧面板的Docs搜索栏中输入“ ee.Image.sampleRegions ”即可。
然后加载训练数据,将80%/20% 用于训练/验证
// 在training要素集中增加一个random属性,值为0到1的随机数
var withRandom = trainging2.randomColumn({columnName:'random',seed:2,distribution: 'uniform',
});var split = 0.8;
var trainingData = withRandom.filter(ee.Filter.lt('random', split));
var validationData = withRandom.filter(ee.Filter.gte('random', split));
以上代码为回归添加了训练和验证数据,
C. 运行 RF 分类器
然后,我们使用训练数据来创建随机森林分类器。尽管我们执行的是回归,而不是分类,这仍然被称为classifier。
// train the RF classification model
var classifier = ee.Classifier.smileRandomForest(100, null, 1, 0.5, null, 0).setOutputMode('REGRESSION').train({features: trainingData, classProperty: 'REDOX_CM', inputProperties: bands});
print(classifier);
注意’ setOutputMode’一定设置为’ REGRESSION '的,该参数对于在 GEE 中运行不同类型的随机森林模型至关重要。
对于随机森林超参数的设置可以查看GEE Docs,描述如下:

最后,现在我们将使用刚刚创建的分类器对图像进行分类。
// Classify the input imagery.
var regression = inStack.classify(classifier, 'predicted');
print(regression);
值得一提的事,在 Earth Engine 中,即使我们正在执行回归任务,但仍然使用的事classify()方法。
到目前为止,我们已经创建了一个空间回归模型,但我们还没有将它添加到我们的地图中,所以如果您运行此代码,您的控制台或地图中不会出现任何新内容(记得顺手ctrl+s)
向地图添加回归结果,创建图例
A. 加载并定义一个连续调色板
由于我们的回归是对连续变量进行分类,因此我们不需要像分类时那样为每个类选择颜色。相反,我们将使用预制的调色板——要访问这些调色板,我们需要加载库。为此,请导入此链接以将模块添加到您的 Reader 存储库
var palettes = require('users/gena/packages:palettes');
如果您想在将来选择不同的连续调色板,请访问此 URL。
现在我们已经加载了所需的库,我们可以为回归输出定义一个调色板。在“选择并定义调色板”的注释下,粘贴:
var palette = palettes.crameri.nuuk[25];
B. 将最终回归结果添加到地图
现在我们已经定义了调色板,我们可以将结果添加到地图中。
// Display the input imagery and the regression classification.
// get dictionaries of min & max predicted value
var regressionMin = (regression.reduceRegion({reducer: ee.Reducer.min(),scale: 100,crs: crs,bestEffort: true,tileScale: 5,geometry: roi
}));
var regressionMax = (regression.reduceRegion({reducer: ee.Reducer.max(),scale: 100,crs: crs,bestEffort: true,tileScale: 5,geometry: roi
}));// Add to map
var viz = {palette: palette,min: regressionMin.getNumber('predicted').getInfo(),max: regressionMax.getNumber('predicted').getInfo()
};
Map.addLayer(regression, viz, 'Regression');
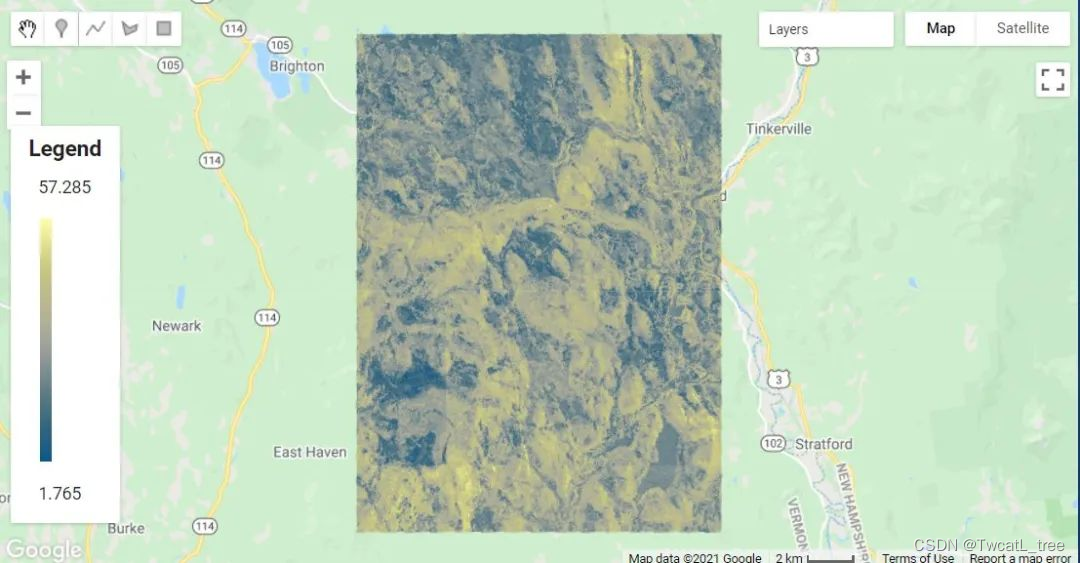
如您所见,在地图上显示回归的结果比显示分类要复杂一些。这是因为该代码的第一部分是为我们的可视化计算适当的最小值和最大值——它只是查找和使用预测氧化还原深度的最高和最低值。在后续计算中,当您使用此代码对不同的连续变量进行建模时,它会自动为您的可视化选择合适的值。
tileScale参数调整用于计算用于在地图上适当显示回归的最小值和最大值的内存。如果您在此练习中遇到内存问题时,可以增加此参数的值。您可以在 GEE 指南中了解有关使用tileScale 和调试的更多信息。
制作图例,将其添加到地图
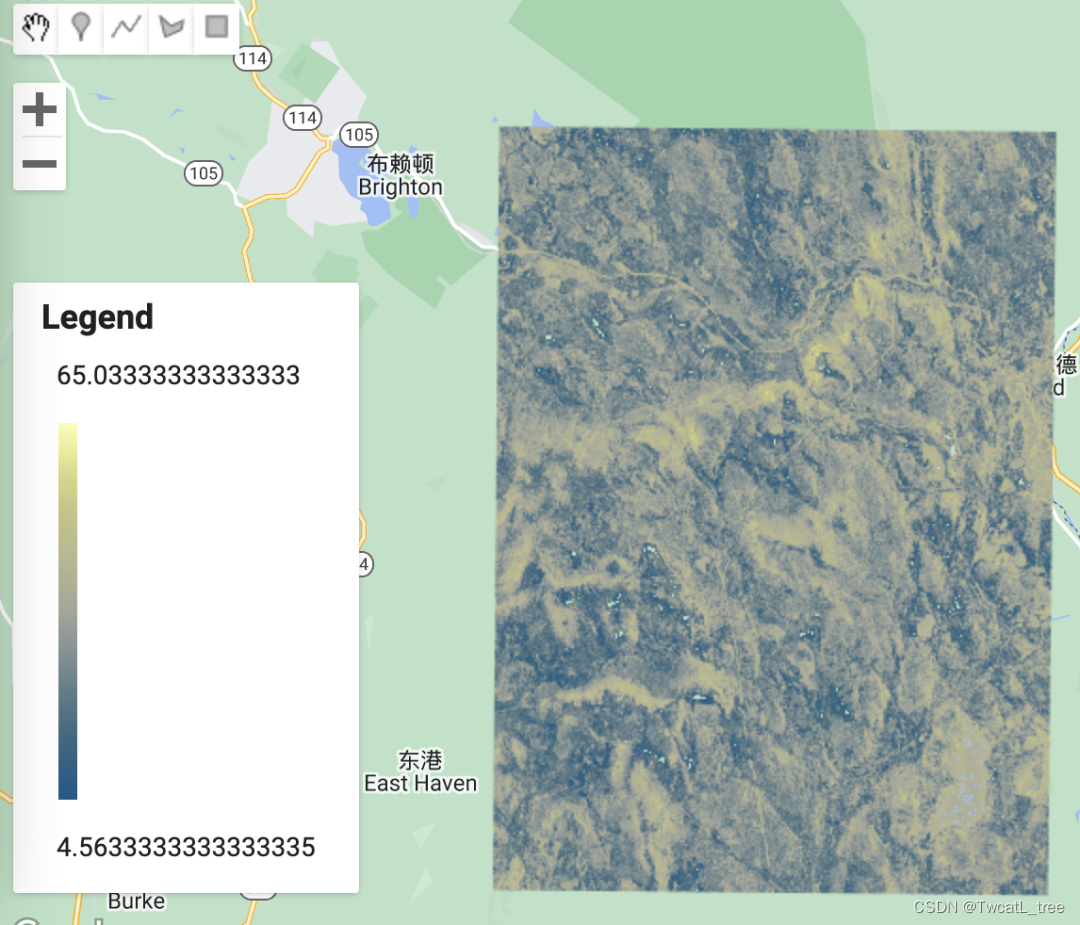
在地图上显示图例总是很有用的,尤其是在处理各种颜色时。
以下代码可能看起来让人头大,但其中大部分只是创建图例的结构和其他美化细节。
// Create the panel for the legend items.
var legend = ui.Panel({style: {position: 'bottom-left',padding: '8px 15px'}
});// Create and add the legend title.
var legendTitle = ui.Label({value: 'Legend',style: {fontWeight: 'bold',fontSize: '18px',margin: '0 0 4px 0',padding: '0'}
});
legend.add(legendTitle);// create the legend image
var lon = ee.Image.pixelLonLat().select('latitude');
var gradient = lon.multiply((viz.max - viz.min) / 100.0).add(viz.min);
var legendImage = gradient.visualize(viz);// create text on top of legend
var panel = ui.Panel({widgets: [ui.Label(viz['max'])],
});legend.add(panel);// create thumbnail from the image
var thumbnail = ui.Thumbnail({image: legendImage,params: {bbox: '0,0,10,100',dimensions: '10x200'},style: {padding: '1px',position: 'bottom-center'}
});// add the thumbnail to the legend
legend.add(thumbnail);// create text on top of legend
var panel = ui.Panel({widgets: [ui.Label(viz['min'])],
});legend.add(panel);
Map.add(legend);// Zoom to the regression on the map
Map.centerObject(roi, 11);
创建模型评估统计数据
可视化评估工具
数据可视化是评估模型性能的一个极其重要的方法,很多时候通过可视化的方式看结果会容易得多。
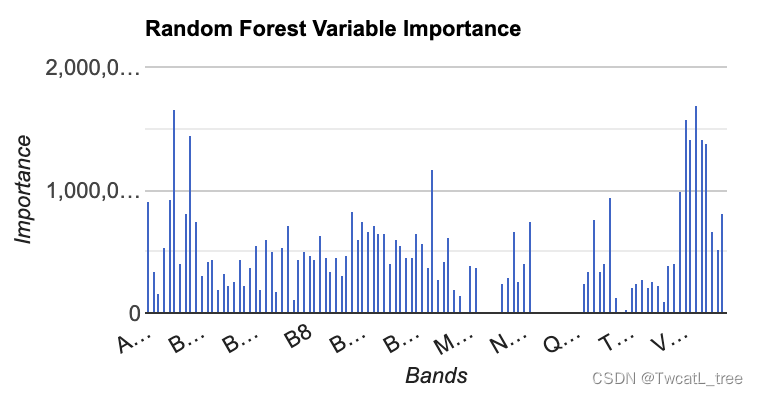
我们要制作的第一个图是具有特征重要性的直方图。这是一个有用的图标,尤其是当我们在模型中使用多个个预测层时。它使我们能够查看哪些变量对模型有帮助,哪些变量可能没有。
// Get variable importance
var dict = classifier.explain();
print("Classifier information:", dict);
var variableImportance = ee.Feature(null, ee.Dictionary(dict).get('importance'));
// Make chart, print it
var chart =ui.Chart.feature.byProperty(variableImportance).setChartType('ColumnChart').setOptions({title: 'Random Forest Variable Importance',legend: {position: 'none'},hAxis: {title: 'Bands'},vAxis: {title: 'Importance'}});
print(chart);
运行代码后,您应该有类似下图所示的内容:
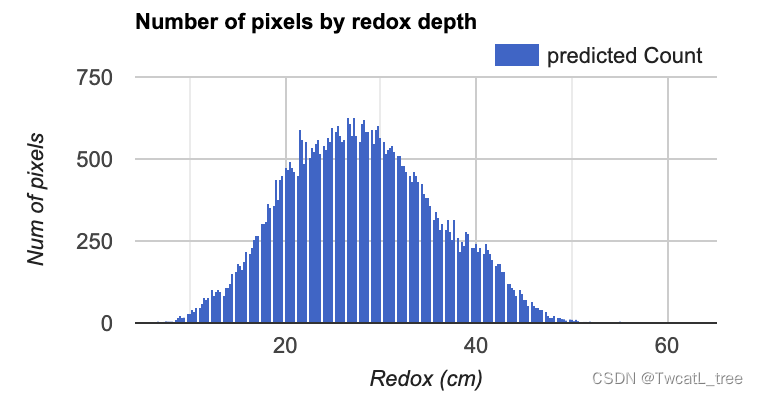
接下来,我们将制作一个直方图,显示在每个氧化还原深度预测我们研究区域中有多少个像素。这是评估预测值分布的有用视觉效果。
// Set chart options
var options = {lineWidth: 1,pointSize: 2,hAxis: {title: 'Redox (cm)'},vAxis: {title: 'Num of pixels'},title: 'Number of pixels by redox depth'
};
var regressionPixelChart = ui.Chart.image.histogram({image: ee.Image(regression),region: roi,scale:100}).setOptions(options);
print(regressionPixelChart);
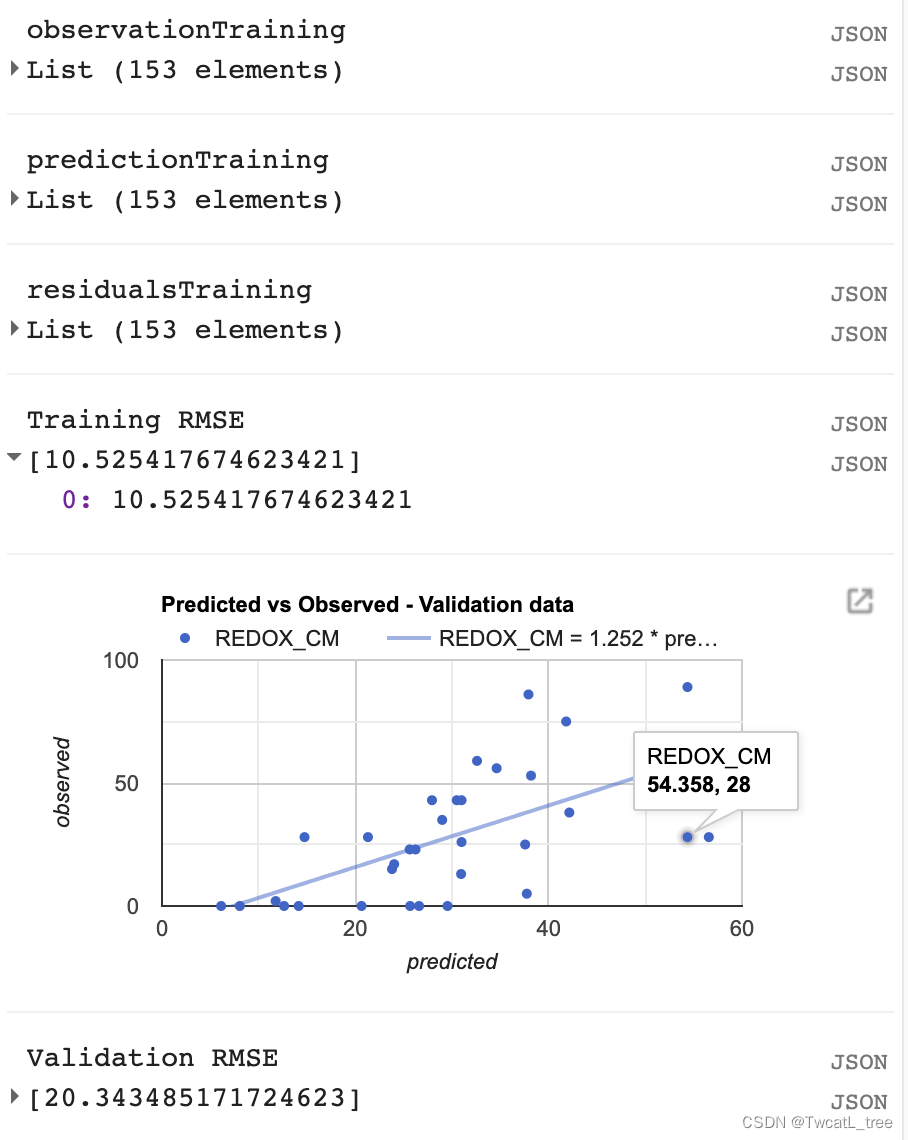
最后,我们将制作的最后一个图是预测值和真实值的散点图。这些对于查看模型的拟合情况十分有帮助,因为它从回归图像(预测值)中获取样本点,并将其与训练数据(真实值)进行对比。
// Get predicted regression points in same location as training data
var predictedTraining = regression.sampleRegions({collection: trainingData,scale:100,geometries: true,projection:crs
});
// Separate the observed (REDOX_CM) and predicted (regression) properties
var sampleTraining = predictedTraining.select(['REDOX_CM', 'predicted']);
// Create chart, print it
var chartTraining = ui.Chart.feature.byFeature({features:sampleTraining, xProperty:'REDOX_CM', yProperties:['predicted']}).setChartType('ScatterChart').setOptions({title: 'Predicted vs Observed - Training data ',hAxis: {'title': 'observed'},vAxis: {'title': 'predicted'},pointSize: 3,trendlines: {0: {showR2: true,visibleInLegend: true},1: {showR2: true,visibleInLegend: true}}});
print(chartTraining);
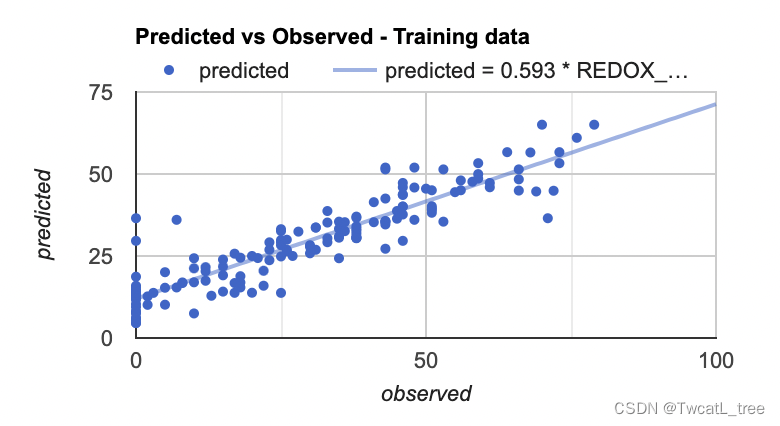
注意:如果您将鼠标悬停在该图的右上角,您也可以看到 R^2 值。(R^2 值会显示在图上)
计算均方根误差 (RMSE)
我们将计算RMSE来评估我们的回归对训练数据的影响。
// Compute RSME
// Get array of observation and prediction values
var observationTraining = ee.Array(sampleTraining.aggregate_array('REDOX_CM'));
var predictionTraining = ee.Array(sampleTraining.aggregate_array('predicted'));
print('observationTraining', observationTraining)
print('predictionTraining', predictionTraining)// Compute residuals
var residualsTraining = observationTraining.subtract(predictionTraining);
print('residualsTraining', residualsTraining)
// Compute RMSE with equation, print it
var rmseTraining = residualsTraining.pow(2).reduce('mean', [0]).sqrt();
print('Training RMSE', rmseTraining);
但是,仅通过查看我们的训练数据,我们无法真正了解我们的数据表现如何。我们将对我们的验证数据执行类似的评估,以了解我们的模型在未用于训练它的数据上的表现如何。
// Get predicted regression points in same location as validation data
var predictedValidation = regression.sampleRegions({collection: validationData,scale:100,geometries: true
});
// Separate the observed (REDOX_CM) and predicted (regression) properties
var sampleValidation = predictedValidation.select(['REDOX_CM', 'predicted']);
// Create chart, print it
var chartValidation = ui.Chart.feature.byFeature(sampleValidation, 'predicted', 'REDOX_CM').setChartType('ScatterChart').setOptions({title: 'Predicted vs Observed - Validation data',hAxis: {'title': 'predicted'},vAxis: {'title': 'observed'},pointSize: 3,trendlines: {0: {showR2: true,visibleInLegend: true},1: {showR2: true,visibleInLegend: true}}});
print(chartValidation);// Get array of observation and prediction values
var observationValidation = ee.Array(sampleValidation.aggregate_array('REDOX_CM'));
var predictionValidation = ee.Array(sampleValidation.aggregate_array('predicted'));
// Compute residuals
var residualsValidation = observationValidation.subtract(predictionValidation);
// Compute RMSE with equation, print it
var rmseValidation = residualsValidation.pow(2).reduce('mean', [0]).sqrt();
print('Validation RMSE', rmseValidation);
导出
// **** Export regression **** //
// create file name for export
var exportName = 'DSM_RF_regression';// If using gmail: Export to Drive
Export.image.toDrive({image: regression,description: exportName,folder: "DigitalSoilMapping",fileNamePrefix: exportName,region: roi,scale: 30,crs: crs,maxPixels: 1e13
});
运行脚本后,窗格右侧的“任务”选项卡将变为橙色,表示有可以运行的导出任务。
单击任务选项卡。
在窗格中找到相应的任务,然后单击“运行”。
在弹出的窗口中,您将看到导出参数。我们已经在代码中指定了这些。请再次检查以确保一切正常,然后单击“运行”。导出可能需要 10 分钟以上才能完成,所以请耐心等待!
请注意,我们正在将导出文件发送到您的 Google Drive 中名为“DigitalSoilMapping”的文件夹中。如果此文件夹尚不存在,则此命令将创建它。
导航到您的 google 驱动器,找到 DigitalSoilMapping 文件夹,然后单击将其打开。
右键单击下载文件,该文件的标题应为“Essex_VT_DSM_regression.tif”。现在,您可以在您选择的 GIS 中打开它。
恭喜!您已成功完成此练习。
// 以下是本次实验的全部代码
var VT_boundary = ee.FeatureCollection("projects/ee-yelu/assets/VT_boundary"),VT_pedons = ee.FeatureCollection("projects/ee-yelu/assets/essex_pedons_all");
// 研究区
var roi = VT_boundary.union();
Map.addLayer(roi, {}, 'shp', false);
var crs = 'EPSG:4326'; // EPSG number for output projection. 32618 = WGS84/UTM Zone 18N. For more info- http://spatialreference.org/ref/epsg/
// S2 影像去云与合成
function maskS2clouds(image) {var qa = image.select('QA60');// Bits 10 and 11 are clouds and cirrus, respectively.var cloudBitMask = 1 << 10;var cirrusBitMask = 1 << 11;// Both flags should be set to zero, indicating clear conditions.var mask = qa.bitwiseAnd(cloudBitMask).eq(0).and(qa.bitwiseAnd(cirrusBitMask).eq(0));return image.updateMask(mask).divide(10000);
}// 计算特征
// NDVI
function NDVI(img) {var ndvi = img.expression("(NIR-R)/(NIR+R)", {"R": img.select("B4"),"NIR": img.select("B8"),});return img.addBands(ndvi.rename("NDVI"));
}// NDWI
function NDWI(img) {var ndwi = img.expression("(G-MIR)/(G+MIR)", {"G": img.select("B3"),"MIR": img.select("B8"),});return img.addBands(ndwi.rename("NDWI"));
}// NDBI
function NDBI(img) {var ndbi = img.expression("(SWIR-NIR)/(SWIR-NIR)", {"NIR": img.select("B8"),"SWIR": img.select("B12"),});return img.addBands(ndbi.rename("NDBI"));
}//SAVI
function SAVI(image) {var savi = image.expression('(NIR - RED) * (1 + 0.5)/(NIR + RED + 0.5)', {'NIR': image.select('B8'),'RED': image.select('B4')}).float();return image.addBands(savi.rename('SAVI'));
}//IBI
function IBI(image) {// Add Index-Based Built-Up Index (IBI)var ibiA = image.expression('2 * SWIR1 / (SWIR1 + NIR)', {'SWIR1': image.select('B6'),'NIR': image.select('B5')}).rename(['IBI_A']);var ibiB = image.expression('(NIR / (NIR + RED)) + (GREEN / (GREEN + SWIR1))', {'NIR': image.select('B8'),'RED': image.select('B4'),'GREEN': image.select('B3'),'SWIR1': image.select('B11')}).rename(['IBI_B']);var ibiAB = ibiA.addBands(ibiB);var ibi = ibiAB.normalizedDifference(['IBI_A', 'IBI_B']);return image.addBands(ibi.rename(['IBI']));
}//RVI
function RVI(image) {var rvi = image.expression('NIR/Red', {'NIR': image.select('B8'),'Red': image.select('B4')});return image.addBands(rvi.rename('RVI'));
}//DVI
function DVI(image) {var dvi = image.expression('NIR - Red', {'NIR': image.select('B8'),'Red': image.select('B4')}).float();return image.addBands(dvi.rename('DVI'));
}// 创建var inStack = ee.Image()
for (var i = 0; i < 3; i += 1) {var start = ee.Date('2019-03-01').advance(30 * i, 'day');print(start)var end = start.advance(30, 'day');var dataset = ee.ImageCollection('COPERNICUS/S2_SR').filterDate(start, end).filterBounds(roi).filter(ee.Filter.lt('CLOUDY_PIXEL_PERCENTAGE',75)).map(maskS2clouds).map(NDVI).map(NDWI).map(NDBI).map(SAVI).map(IBI).map(RVI).map(DVI);var inStack_monthly = dataset.median().clip(roi);// 可视化var visualization = {min: 0.0,max: 0.3,bands: ['B4', 'B3', 'B2'],};Map.addLayer(inStack_monthly, visualization, 'S2_' + i, false);// // 纹理特征var B8 = inStack_monthly.select('B8').multiply(100).toInt16();var glcm = B8.glcmTexture({size: 3});var contrast = glcm.select('B8_contrast');var var_ = glcm.select('B8_var');var savg = glcm.select('B8_savg');var dvar = glcm.select('B8_dvar');inStack_monthly = inStack_monthly.addBands([contrast, var_, savg, dvar])// Sentinel-1 var imgVV = ee.ImageCollection('COPERNICUS/S1_GRD').filter(ee.Filter.listContains('transmitterReceiverPolarisation', 'VV')).filter(ee.Filter.listContains('transmitterReceiverPolarisation', 'VH')).filter(ee.Filter.eq('instrumentMode', 'IW')).filterBounds(roi).map(function(image) {var edge = image.lt(-30.0);var maskedImage = image.mask().and(edge.not());return image.updateMask(maskedImage);});var img_S1_asc = imgVV.filter(ee.Filter.eq('orbitProperties_pass', 'ASCENDING'));var VV_img = img_S1_asc.filterDate(start, end).select("VV").median().clip(roi);var VH_img = img_S1_asc.filterDate(start, end).select("VH").median().clip(roi);inStack_monthly = inStack_monthly.addBands([VV_img, VH_img])print("img_S2_monthly", inStack_monthly)inStack = inStack.addBands(inStack_monthly.select(inStack_monthly.bandNames()))
}
inStack = inStack.select(inStack.bandNames().remove("constant"))
print(inStack)
inStack.reproject(crs, null, 30);
Map.centerObject(inStack)var trainingFeatureCollection = ee.FeatureCollection(VT_pedons, 'geometry');
print(bands)
var training = inStack.reduceRegions({collection:trainingFeatureCollection,reducer :ee.Reducer.mean(),scale:100,tileScale :5,crs: crs
});
print(training)
var bands = inStack.bandNames()var trainging2 = training.filter(ee.Filter.notNull(bands));
// print(trainging2);// 在training要素集中增加一个random属性,值为0到1的随机数
var withRandom = trainging2.randomColumn({columnName:'random',seed:2,distribution: 'uniform',
});var split = 0.8;
var trainingData = withRandom.filter(ee.Filter.lt('random', split));
var validationData = withRandom.filter(ee.Filter.gte('random', split)); // train the RF classification model
var classifier = ee.Classifier.smileRandomForest(100, null, 1, 0.5, null, 0).setOutputMode('REGRESSION').train({features: trainingData, classProperty: 'REDOX_CM', inputProperties: bands});
print(classifier);// Classify the input imagery.
var regression = inStack.classify(classifier, 'predicted');
print(regression);var palettes = require('users/gena/packages:palettes');
var palette = palettes.crameri.nuuk[25];
print("regression", regression)
// Display the input imagery and the regression classification.
// get dictionaries of min & max predicted value
var regressionMin = (regression.reduceRegion({reducer: ee.Reducer.min(),scale: 100,crs: crs,bestEffort: true,tileScale: 5,geometry: roi
}));
var regressionMax = (regression.reduceRegion({reducer: ee.Reducer.max(),scale: 100,crs: crs,bestEffort: true,tileScale: 5,geometry: roi
}));// Add to map
var viz = {palette: palette,min: regressionMin.getNumber('predicted').getInfo(),max: regressionMax.getNumber('predicted').getInfo()
};
Map.addLayer(regression, viz, 'Regression');// Create the panel for the legend items.
var legend = ui.Panel({style: {position: 'bottom-left',padding: '8px 15px'}
});// Create and add the legend title.
var legendTitle = ui.Label({value: 'Legend',style: {fontWeight: 'bold',fontSize: '18px',margin: '0 0 4px 0',padding: '0'}
});
legend.add(legendTitle);// create the legend image
var lon = ee.Image.pixelLonLat().select('latitude');
var gradient = lon.multiply((viz.max - viz.min) / 100.0).add(viz.min);
var legendImage = gradient.visualize(viz);// create text on top of legend
var panel = ui.Panel({widgets: [ui.Label(viz['max'])],
});legend.add(panel);// create thumbnail from the image
var thumbnail = ui.Thumbnail({image: legendImage,params: {bbox: '0,0,10,100',dimensions: '10x200'},style: {padding: '1px',position: 'bottom-center'}
});// add the thumbnail to the legend
legend.add(thumbnail);// create text on top of legend
var panel = ui.Panel({widgets: [ui.Label(viz['min'])],
});legend.add(panel);
Map.add(legend);// Zoom to the regression on the map
Map.centerObject(roi, 11);// Get variable importance
var dict = classifier.explain();
print("Classifier information:", dict);
var variableImportance = ee.Feature(null, ee.Dictionary(dict).get('importance'));
// Make chart, print it
var chart =ui.Chart.feature.byProperty(variableImportance).setChartType('ColumnChart').setOptions({title: 'Random Forest Variable Importance',legend: {position: 'none'},hAxis: {title: 'Bands'},vAxis: {title: 'Importance'}});
print(chart);// Set chart options
var options = {lineWidth: 1,pointSize: 2,hAxis: {title: 'Redox (cm)'},vAxis: {title: 'Num of pixels'},title: 'Number of pixels by redox depth'
};
var regressionPixelChart = ui.Chart.image.histogram({image: ee.Image(regression),region: roi,scale:100}).setOptions(options);
print(regressionPixelChart);// Get predicted regression points in same location as training data
var predictedTraining = regression.sampleRegions({collection: trainingData,scale:100,geometries: true,projection:crs
});
// Separate the observed (REDOX_CM) and predicted (regression) properties
var sampleTraining = predictedTraining.select(['REDOX_CM', 'predicted']);
// Create chart, print it
var chartTraining = ui.Chart.feature.byFeature({features:sampleTraining, xProperty:'REDOX_CM', yProperties:['predicted']}).setChartType('ScatterChart').setOptions({title: 'Predicted vs Observed - Training data ',hAxis: {'title': 'observed'},vAxis: {'title': 'predicted'},pointSize: 3,trendlines: {0: {showR2: true,visibleInLegend: true},1: {showR2: true,visibleInLegend: true}}});
print(chartTraining);// **** Compute RSME ****
// Get array of observation and prediction values
var observationTraining = ee.Array(sampleTraining.aggregate_array('REDOX_CM'));
var predictionTraining = ee.Array(sampleTraining.aggregate_array('predicted'));
print('observationTraining', observationTraining)
print('predictionTraining', predictionTraining)// Compute residuals
var residualsTraining = observationTraining.subtract(predictionTraining);
print('residualsTraining', residualsTraining)
// Compute RMSE with equation, print it
var rmseTraining = residualsTraining.pow(2).reduce('mean', [0]).sqrt();
print('Training RMSE', rmseTraining);// Get predicted regression points in same location as validation data
var predictedValidation = regression.sampleRegions({collection: validationData,scale:100,geometries: true
});
// Separate the observed (REDOX_CM) and predicted (regression) properties
var sampleValidation = predictedValidation.select(['REDOX_CM', 'predicted']);
// Create chart, print it
var chartValidation = ui.Chart.feature.byFeature(sampleValidation, 'predicted', 'REDOX_CM').setChartType('ScatterChart').setOptions({title: 'Predicted vs Observed - Validation data',hAxis: {'title': 'predicted'},vAxis: {'title': 'observed'},pointSize: 3,trendlines: {0: {showR2: true,visibleInLegend: true},1: {showR2: true,visibleInLegend: true}}});
print(chartValidation);// Get array of observation and prediction values
var observationValidation = ee.Array(sampleValidation.aggregate_array('REDOX_CM'));
var predictionValidation = ee.Array(sampleValidation.aggregate_array('predicted'));
// Compute residuals
var residualsValidation = observationValidation.subtract(predictionValidation);
// Compute RMSE with equation, print it
var rmseValidation = residualsValidation.pow(2).reduce('mean', [0]).sqrt();
print('Validation RMSE', rmseValidation);// **** Export regression **** //
// create file name for export
var exportName = 'DSM_RF_regression';// If using gmail: Export to Drive
Export.image.toDrive({image: regression,description: exportName,folder: "DigitalSoilMapping",fileNamePrefix: exportName,region: roi,scale: 30,crs: crs,maxPixels: 1e13
});