######--------ssGSEA---------##############
#加载18条神经通路合集
load("./score/ssGSEA/geneset_list_18.RData")
geneset_list
select_pathway_name <- c("G_protein_coupled_ACh_receptor_signalling","Response_to_ACh","ACh_receptor_binding")#个性化修改# ssGSEA计算
expression_matrix <- as.matrix(MERGE@assays$RNA@data)# 提取表达矩阵ssgsea_scores <- gsva(expr = expression_matrix, gset.idx.list = geneset_list[select_pathway_name], method = "ssgsea", kcdf = "Gaussian", abs.ranking = TRUE
)ssgsea_scores[1:3,1:4]# 将ssGSEA分数添加到 Seurat 对象
ssgsea_scores <- t(ssgsea_scores) # 转置,使每行对应细胞,每列对应基因集
MERGE <- AddMetaData(MERGE, metadata = as.data.frame(ssgsea_scores))
head(MERGE@meta.data)# 画图
# 1.山脊图
score_columns <- c("G_protein_coupled_ACh_receptor_signalling","Response_to_ACh","ACh_receptor_binding")for (score_name in score_columns) {p <- ggplot(MERGE@meta.data, aes_string(x = paste0(score_name), y = "cluster", fill = "cluster")) +geom_density_ridges(scale = 1.5, alpha = 0.7, color = "white") +scale_fill_manual(values = color_epi) +theme_bw() +theme(axis.text.x = element_text(size = 12, face = "plain"),axis.text.y = element_text(size = 12, face = "plain"),axis.title.x = element_blank(), # 移除x轴标题axis.title.y = element_text(size = 14, face = "plain"),plot.title = element_text(size = 16, face = "bold", hjust = 0.5), # 设置标题样式panel.grid.major = element_blank(),panel.grid.minor = element_blank(),legend.position = "none") +labs(title = paste0(score_name, " Score"), # 主标题y = "", fill = NULL)# 保存图片ggsave(paste0("ssGSEA_", score_name, "_ridge.png"), p, height = 4, width = 7)
}
ssGSEA-山脊图-汇总18条神经通路合集
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如若转载,请注明出处:http://www.hqwc.cn/news/906354.html
如若内容造成侵权/违法违规/事实不符,请联系编程知识网进行投诉反馈email:809451989@qq.com,一经查实,立即删除!相关文章
Django 之 ContentType(django_content_type)
REF
https://blog.csdn.net/weixin_30498807/article/details/96845973
https://blog.csdn.net/aaronthon/article/details/81714496用来管理和维护我们应用程序的models转自:https://www.cnblogs.com/oysq/p/15643370.htmlDjango除了我们常见的admin、auth、session等contrib…
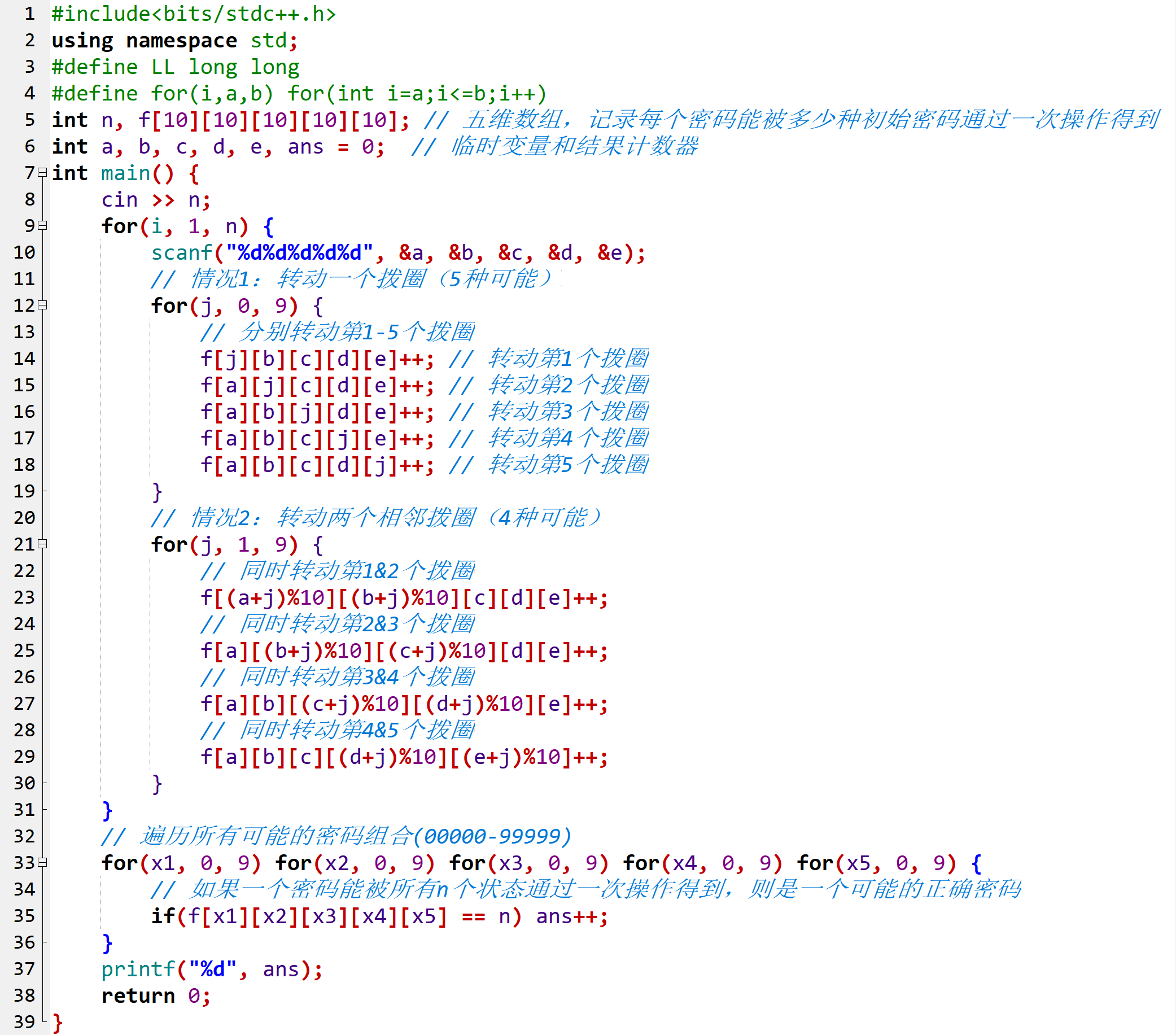
20243402谢子凌 实验二《Python程序设计》实验报告
学号 2024-2025-2 《Python程序设计》实验二报告
课程:《Python程序设计》
班级: 2434
姓名: 谢子凌
学号:20243402
实验教师:王志强老师
实验日期:2025年3月26日
必修/选修: 公选课
1.实验内容
设计并完成一个完整的应用程序,完成加减乘除模等运算,功能多多益善。
考…
【视频】文本挖掘专题:Python、R用LSTM情感语义分析实例合集|上市银行年报、微博评论、红楼梦数据、汽车口碑数据采集词云可视化
原文链接:https://tecdat.cn/?p=41149原文出处:拓端数据部落公众号
分析师:Zhenzhen Liu,Shuai FungPython企业年报文本分析情感挖掘语调分析:以上市银行为例作为数据科学家,我们始终关注如何从非结构化数据中提取高价值信息。本专题合集聚焦企业年报的文本分析技术,通…
20242125 单嘉怡 实验二《Python程序设计》实验报告
20242125单嘉怡 2024-2025-2 《Python程序设计》实验二报告
课程:《Python程序设计》
班级:2421
姓名:单嘉怡
学号:20232401
实验教师:王志强
实验日期:2025年3月26日
必修/选修: 公选课
一.实验内容设计并完成一个完整的应用程序,完成加减乘除模等运算,功能多多益善。…
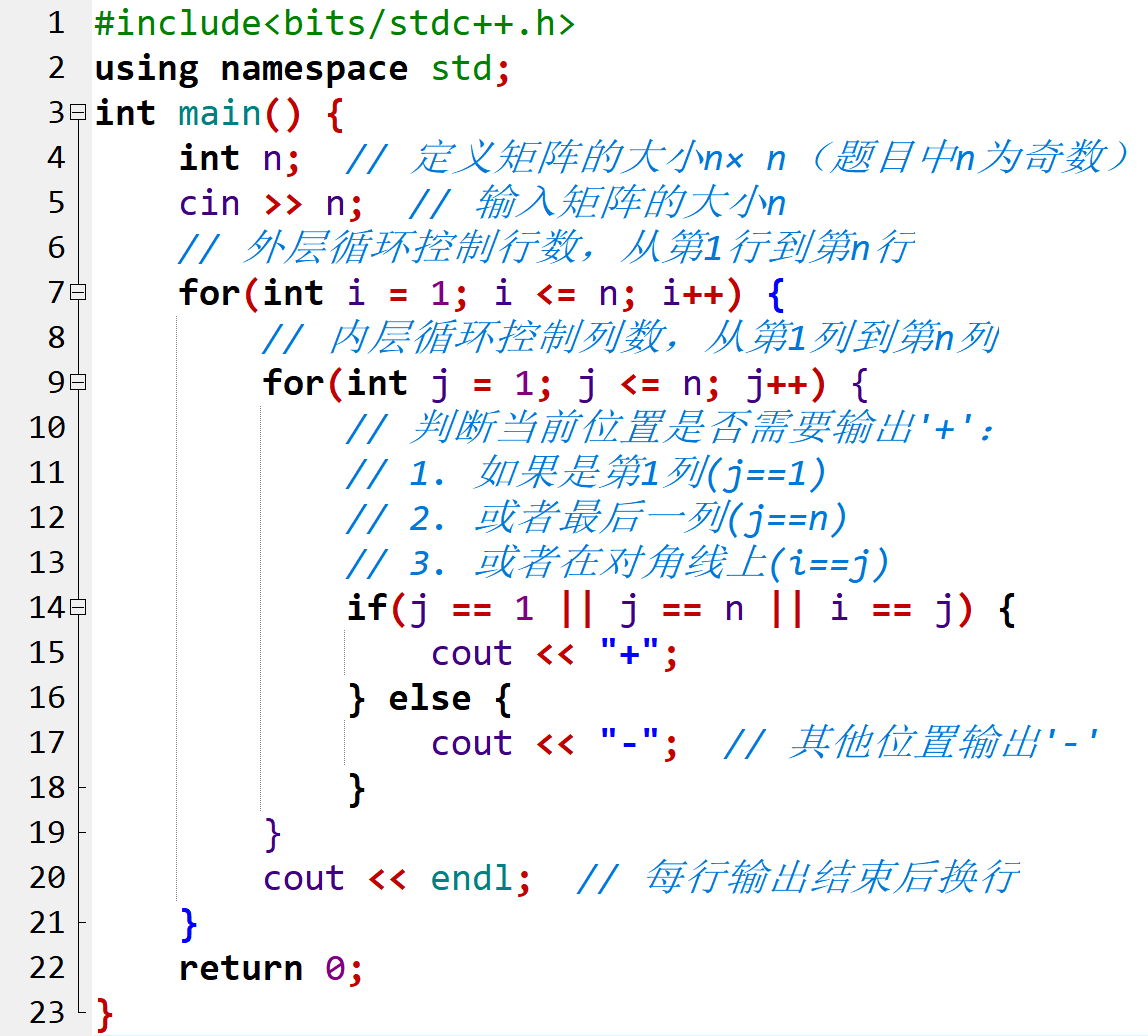
20243116 实验二《Python程序设计》实验报告
20243116 2024-2025-2 《Python程序设计》实验二报告
课程:《Python程序设计》
班级: 2431
姓名: 陆翔轩
学号:20243116
实验教师:王志强
实验日期:2025年3月26日
必修/选修: 公选课
一、实验内容
1.设计并完成一个完整的应用程序,完成加减乘除模等运算,功能多多益善。…
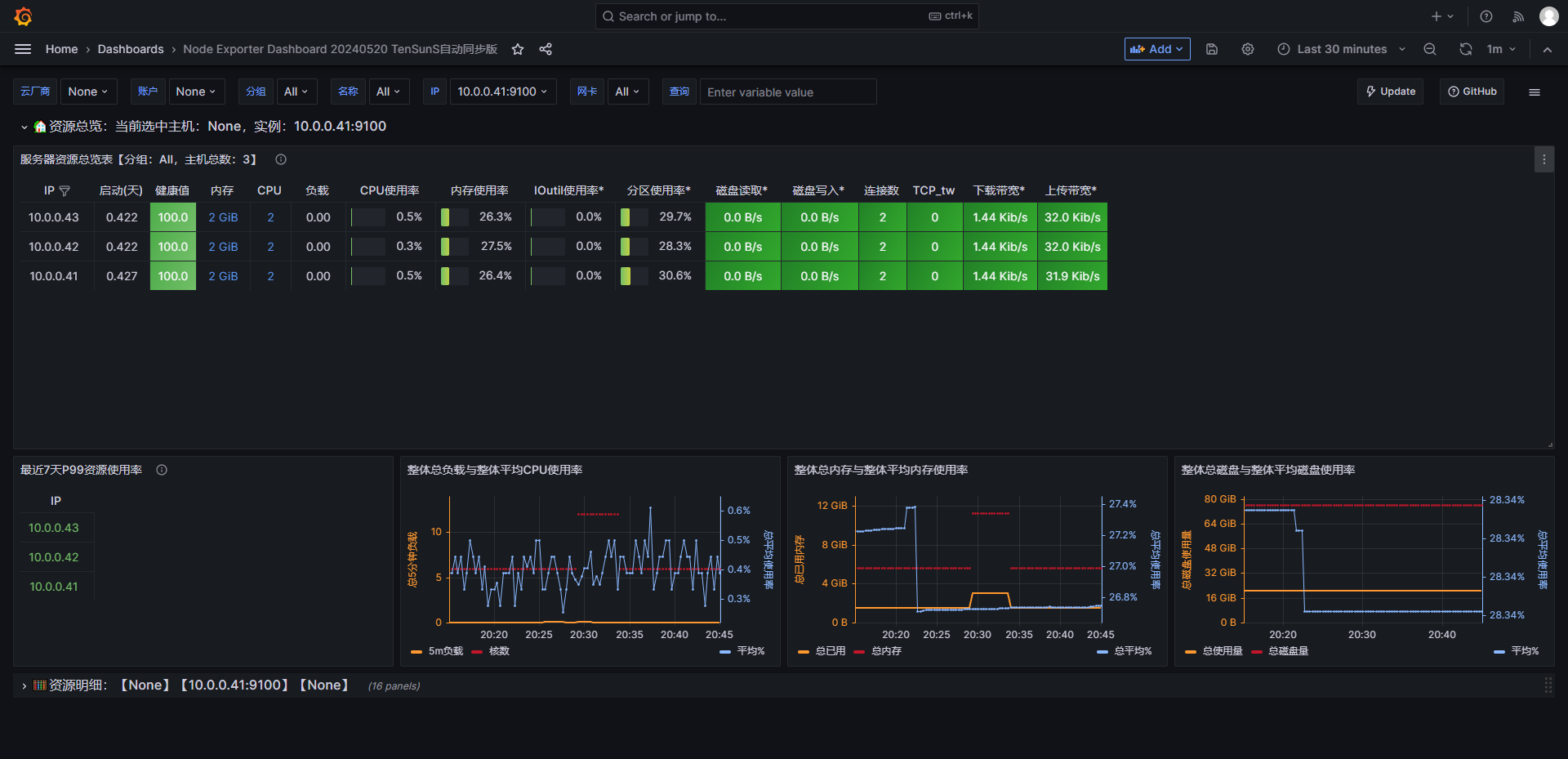
基于Grafana使用官方模版查询Prometheus数据
grafana配置Prometheus作为数据源选择一个模版,导入到dashboards
模板地址
https://grafana.com/grafana/dashboards/
选择Prometheus,选择一个适应版本的模版,记录ID
例如:1860 11074 8919查看dashboards