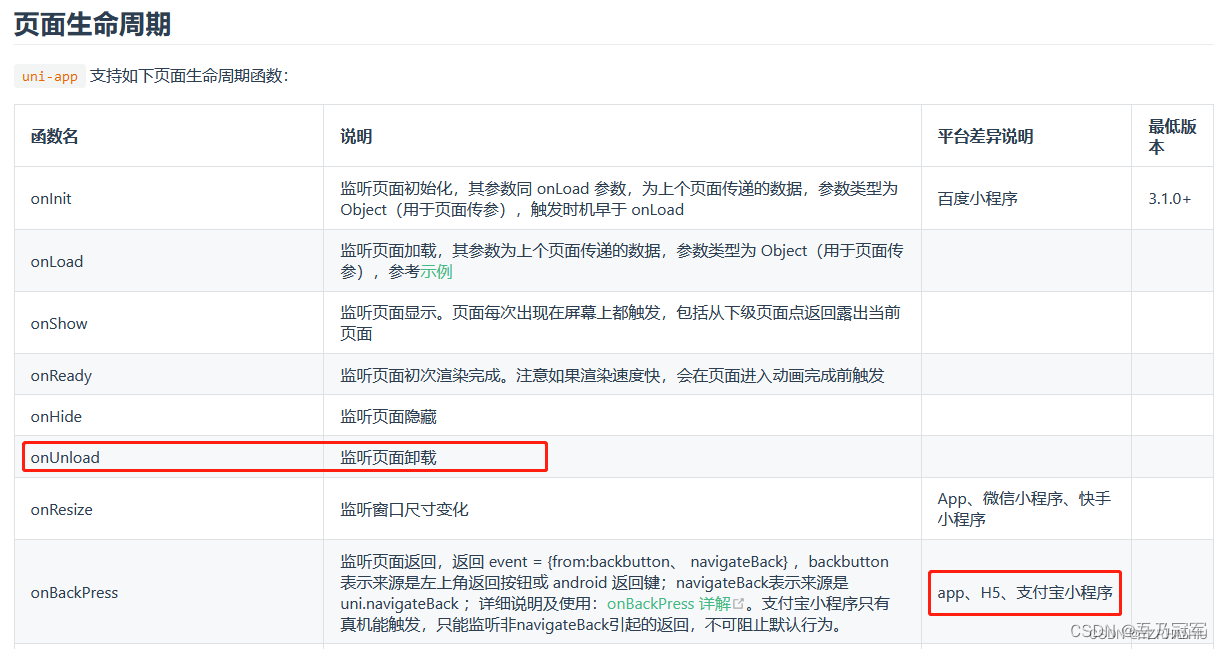
遗传算法的核心,就在于,把待求的变量转化成二进制串,二进制串就像dna,可以对它的其中某几位进行交换,变异等操作,然后再转换回十进制,带入目标函数,计算适应度,保留适应度高变量进行繁殖。最关键的地方就是十进制数和二进制之间的相互编码与解码。
举个例子,如果我们有一个待求的变量x,取值范围是的float型,我们可以把它编码成一个10位的二进制串:
首先把的区间映射到
的区间,
再把转成二进制串:10位二进制串可以表示的最大十进制数是
,将
乘
,
,再四舍五入取整,就得到了0到1023之间的整数,假如
,65转化成十位二进制串就是 0 0 0 1 0 0 0 0 0 1,对这个二进制串就可以很容易的进行交换和变异操作了。反之,逆过程就是dna到十进制变量的解码。
如果有2个未知数,dna就是20位二进制串,前十位是第一个变量,后十位是第二个变量,以此类推。也可以奇数位是第一个变量,偶数位是第二个变量,可以自己定义编码解码方式。
完整python代码
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import cm
from mpl_toolkits.mplot3d import Axes3DDNA_SIZE = 24
POP_SIZE = 80
CROSSOVER_RATE = 0.6
MUTATION_RATE = 0.01
N_GENERATIONS = 100
X_BOUND = [-2.048, 2.048]
Y_BOUND = [-2.048, 2.048]def F(x, y):return 100.0 * (y - x ** 2.0) ** 2.0 + (1 - x) ** 2.0 # 以香蕉函数为例def plot_3d(ax):X = np.linspace(*X_BOUND, 100)Y = np.linspace(*Y_BOUND, 100)X, Y = np.meshgrid(X, Y)Z = F(X, Y)ax.plot_surface(X, Y, Z, rstride=1, cstride=1, cmap=cm.coolwarm)ax.set_xlabel('x')ax.set_ylabel('y')ax.set_zlabel('z')plt.pause(3)plt.show()def get_fitness(pop):x, y = translateDNA(pop)pred = F(x, y)return pred# return pred - np.min(pred)+1e-3 # 求最大值时的适应度# return np.max(pred) - pred + 1e-3 # 求最小值时的适应度,通过这一步fitness的范围为[0, np.max(pred)-np.min(pred)]def translateDNA(pop): # pop表示种群矩阵,一行表示一个二进制编码表示的DNA,矩阵的行数为种群数目x_pop = pop[:, 0:DNA_SIZE] # 前DNA_SIZE位表示Xy_pop = pop[:, DNA_SIZE:] # 后DNA_SIZE位表示Yx = x_pop.dot(2 ** np.arange(DNA_SIZE)[::-1]) / float(2 ** DNA_SIZE - 1) * (X_BOUND[1] - X_BOUND[0]) + X_BOUND[0]y = y_pop.dot(2 ** np.arange(DNA_SIZE)[::-1]) / float(2 ** DNA_SIZE - 1) * (Y_BOUND[1] - Y_BOUND[0]) + Y_BOUND[0]return x, ydef crossover_and_mutation(pop, CROSSOVER_RATE=0.8):new_pop = []for father in pop: # 遍历种群中的每一个个体,将该个体作为父亲child = father # 孩子先得到父亲的全部基因(这里我把一串二进制串的那些0,1称为基因)if np.random.rand() < CROSSOVER_RATE: # 产生子代时不是必然发生交叉,而是以一定的概率发生交叉mother = pop[np.random.randint(POP_SIZE)] # 再种群中选择另一个个体,并将该个体作为母亲cross_points = np.random.randint(low=0, high=DNA_SIZE * 2) # 随机产生交叉的点child[cross_points:] = mother[cross_points:] # 孩子得到位于交叉点后的母亲的基因mutation(child) # 每个后代有一定的机率发生变异new_pop.append(child)return new_popdef mutation(child, MUTATION_RATE=0.003):if np.random.rand() < MUTATION_RATE: # 以MUTATION_RATE的概率进行变异mutate_point = np.random.randint(0, DNA_SIZE*2) # 随机产生一个实数,代表要变异基因的位置child[mutate_point] = child[mutate_point] ^ 1 # 将变异点的二进制为反转def select(pop, fitness): # nature selection wrt pop's fitnessidx = np.random.choice(np.arange(POP_SIZE), size=POP_SIZE, replace=True,p=(fitness) / (fitness.sum()))return pop[idx]def print_info(pop):fitness = get_fitness(pop)max_fitness_index = np.argmax(fitness)print("max_fitness:", fitness[max_fitness_index])x, y = translateDNA(pop)print("最优的基因型:", pop[max_fitness_index])print("(x, y):", (x[max_fitness_index], y[max_fitness_index]))print(F(x[max_fitness_index], y[max_fitness_index]))if __name__ == "__main__":fig = plt.figure()ax = Axes3D(fig)plt.ion() # 将画图模式改为交互模式,程序遇到plt.show不会暂停,而是继续执行plot_3d(ax)pop = np.random.randint(2, size=(POP_SIZE, DNA_SIZE * 2)) # matrix (POP_SIZE, DNA_SIZE)for _ in range(N_GENERATIONS): # 迭代N代x, y = translateDNA(pop)if 'sca' in locals():sca.remove()sca = ax.scatter(x, y, F(x, y), c='black', marker='o')plt.show()plt.pause(0.1)pop = np.array(crossover_and_mutation(pop, CROSSOVER_RATE))fitness = get_fitness(pop)pop = select(pop, fitness) # 选择生成新的种群print_info(pop)plt.ioff()plot_3d(ax)

![[NISACTF 2022]checkin](https://img-blog.csdnimg.cn/img_convert/1892ac0aa5205deedf990641435e0d98.png)