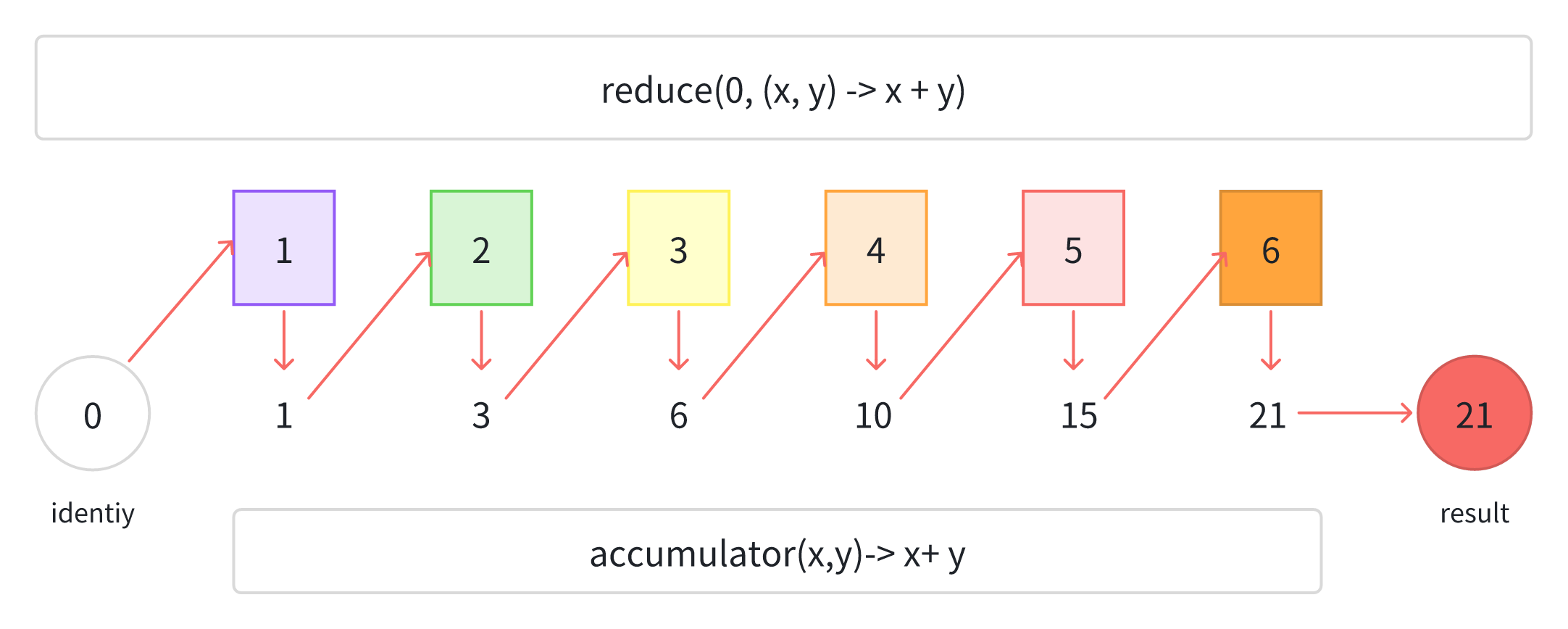
问题:给Taxonomy排序,并筛选OTU表中存在的

根据提示运行:rlang::last_error(),显示:
Backtrace:▆1. ├─dplyr::arrange(taxonomy, phylum, class, order, family, full)2. ├─dplyr:::arrange.data.frame(taxonomy, phylum, class, order, family, full)3. │ └─dplyr:::arrange_rows(.data, dots = dots, locale = .locale)4. │ ├─dplyr::mutate(data, `:=`("{name}", !!dot), .keep = "none")5. │ └─dplyr:::mutate.data.frame(data, `:=`("{name}", !!dot), .keep = "none")6. │ └─dplyr:::mutate_cols(.data, dplyr_quosures(...), by)7. │ ├─base::withCallingHandlers(...)8. │ └─dplyr:::mutate_col(dots[[i]], data, mask, new_columns)9. │ └─mask$eval_all_mutate(quo)10. │ └─dplyr (local) eval()11. └─base::.handleSimpleError(`<fn>`, "找不到对象'phylum'", base::quote(NULL))12. └─dplyr (local) h(simpleError(msg, call))13. └─rlang::abort(message, class = error_class, parent = parent, call = error_call)
最后发现,是数据中的“phylum”大小写不对,导致找不到对象,一一对应后即可顺利推进。