#2.2挑选term---selected_clusterenrich=enrichmets[grepl(pattern = "cilium|matrix|excular|BMP|inflamm|development|muscle|vaso|pulmonary|alveoli",x = enrichmets$Description),]head(selected_clusterenrich) distinct(selected_clusterenrich)# remove duplicate rows based on Description 并且保留其他所有变量
distinct_df <- distinct(enrichmets, Description,.keep_all = TRUE)library(ggplot2)
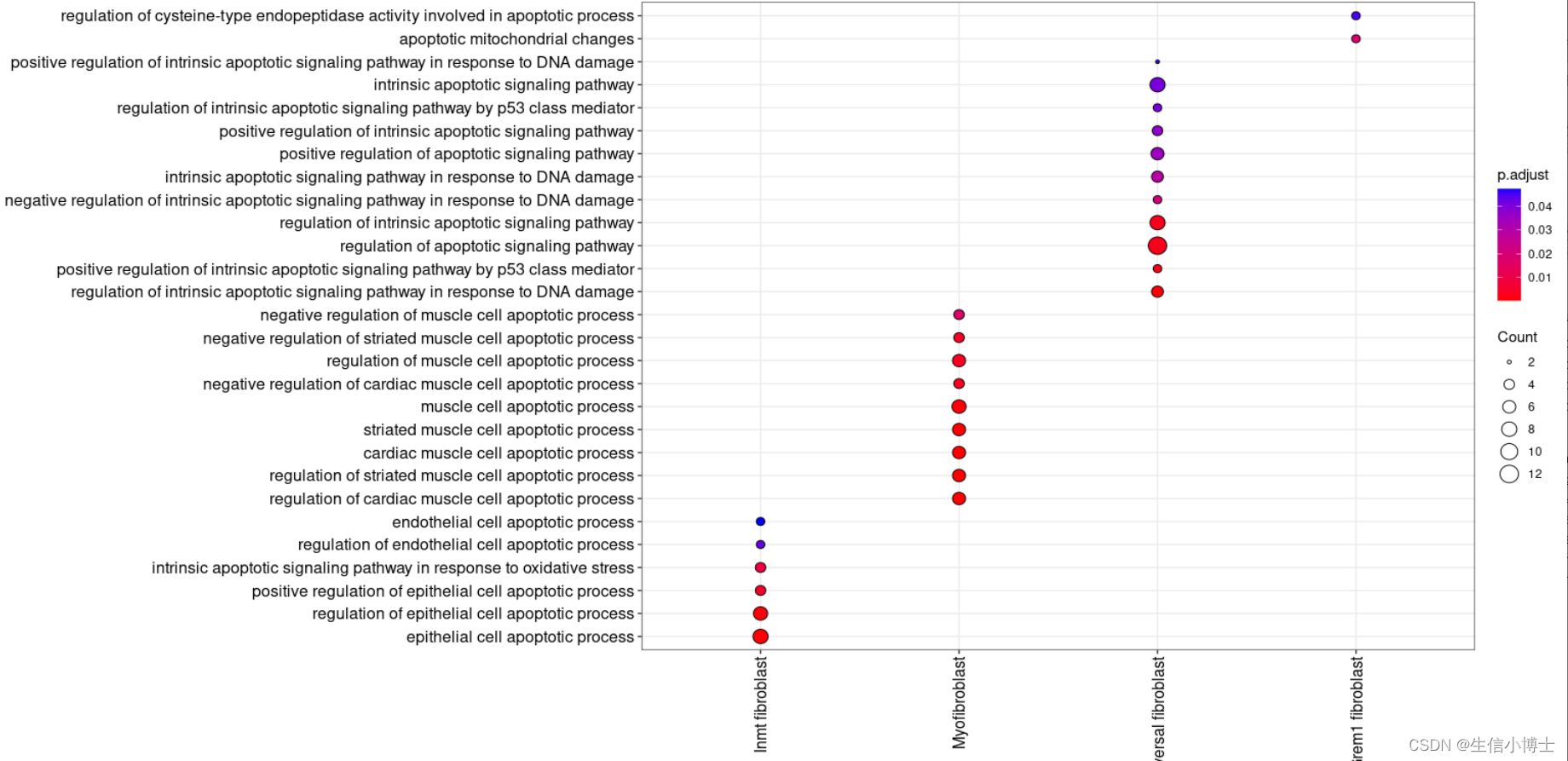
ggplot( distinct_df %>%dplyr::filter(stringr::str_detect(pattern = "cilium|matrix|excular|BMP|inflamm|development|muscle",Description)) %>%group_by(Description) %>%add_count() %>%dplyr::arrange(dplyr::desc(n),dplyr::desc(Description)) %>%mutate(Description =forcats:: fct_inorder(Description)), #fibri|matrix|collaaes(Cluster, Description)) +geom_point(aes(fill=p.adjust, size=Count), shape=21)+theme_bw()+theme(axis.text.x=element_text(angle=90,hjust = 1,vjust=0.5),axis.text.y=element_text(size = 12),axis.text = element_text(color = 'black', size = 12))+scale_fill_gradient(low="red",high="blue")+labs(x=NULL,y=NULL)
# coord_flip()head(enrichmets)ggplot( distinct(enrichmets,Description,.keep_all=TRUE) %>% # dplyr::mutate(Cluster = factor(Cluster, levels = unique(.$Cluster))) %>%dplyr::mutate(Description = factor(Description, levels = unique(.$Description))) %>%# dplyr::group_by(Cluster) %>%dplyr::filter(stringr::str_detect(pattern = "cilium organization|motile cilium|cilium movemen|cilium assembly|cell-matrix adhesion|extracellular matrix organization|regulation of acute inflammatory response to antigenic stimulus|collagen-containing extracellular matrix|negative regulation of BMP signaling pathway|extracellular matrix structural constituent|extracellular matrix binding|fibroblast proliferation|collagen biosynthetic process|collagen trimer|fibrillar collagen trimer|inflammatory response to antigenic stimulus|chemokine activity|chemokine production|cell chemotaxis|chemoattractant activity|NLRP3 inflammasome complex assembly|inflammatory response to wounding|Wnt signaling pathway|response to oxidative stress|regulation of vascular associated smooth muscle cell proliferation|venous blood vessel development|regulation of developmental growth|lung alveolus development|myofibril assembly|blood vessel diameter maintenance|gas transport|cell maturation|regionalization|oxygen carrier activity|oxygen binding|vascular associated smooth muscle cell proliferation",Description)) %>%# group_by(Description) %>%add_count() %>%dplyr::arrange(dplyr::desc(n),dplyr::desc(Description)) %>%mutate(Description =forcats:: fct_inorder(Description)), #fibri|matrix|collaaes(Cluster, y = Description)) + #stringr:: str_wrapgeom_point(aes(fill=p.adjust, size=Count), shape=21)+theme_bw()+theme(axis.text.x=element_text(angle=90,hjust = 1,vjust=0.5),axis.text.y=element_text(size = 12),axis.text = element_text(color = 'black', size = 12))+scale_fill_gradient(low="red",high="blue")+labs(x=NULL,y=NULL)
# coord_flip()
print(getwd())p=ggplot( distinct(enrichmets,Description,.keep_all=TRUE) %>%dplyr::mutate(Description = factor(Description, levels = unique(.$Description))) %>% #调整terms显示顺序dplyr::filter(stringr::str_detect(pattern = "cilium organization|motile cilium|cilium movemen|cilium assembly|cell-matrix adhesion|extracellular matrix organization|regulation of acute inflammatory response to antigenic stimulus|collagen-containing extracellular matrix|negative regulation of BMP signaling pathway|extracellular matrix structural constituent|extracellular matrix binding|fibroblast proliferation|collagen biosynthetic process|collagen trimer|fibrillar collagen trimer|inflammatory response to antigenic stimulus|chemokine activity|chemokine production|cell chemotaxis|chemoattractant activity|NLRP3 inflammasome complex assembly|inflammatory response to wounding|Wnt signaling pathway|response to oxidative stress|regulation of vascular associated smooth muscle cell proliferation|venous blood vessel development|regulation of developmental growth|lung alveolus development|myofibril assembly|blood vessel diameter maintenance|gas transport|cell maturation|regionalization|oxygen carrier activity|oxygen binding|vascular associated smooth muscle cell proliferation",Description)) %>%group_by(Description) %>%add_count() %>%dplyr::arrange(dplyr::desc(n),dplyr::desc(Description)) %>%mutate(Description =forcats:: fct_inorder(Description)), #fibri|matrix|collaaes(Cluster, y = Description)) + #stringr:: str_wrap#scale_y_discrete(labels = function(x) stringr::str_wrap(x, width = 60)) + #调整terms长度geom_point(aes(fill=p.adjust, size=Count), shape=21)+theme_bw()+theme(axis.text.x=element_text(angle=90,hjust = 1,vjust=0.5),axis.text.y=element_text(size = 12),axis.text = element_text(color = 'black', size = 12))+scale_fill_gradient(low="red",high="blue")+labs(x=NULL,y=NULL)
# coord_flip()
print(getwd())ggsave(filename ="~/silicosis/spatial/sp_cluster_rigions_after_harmony/enrichents12.pdf",plot = p,width = 10,height = 12,limitsize = FALSE)######展示term内所有基因,用热图展示-------#提取画图的数据p$data#提取图形中的所有基因-----
mygenes= p$data $geneID %>% stringr::str_split(.,"/",simplify = TRUE) %>%as.vector() %>%unique()
frame_for_genes=p$data %>%as.data.frame() %>% dplyr::group_by(Cluster) #后面使用split的话,必须按照分组排序
head(frame_for_genes)my_genelist= split(frame_for_genes, frame_for_genes$Cluster, drop = TRUE) %>% #注意drop参数的理解lapply(function(x) select(x, geneID));my_genelistmy_genelist= split(frame_for_genes, frame_for_genes$Cluster, drop = TRUE) %>% #注意drop参数的理解lapply(function(x) x$geneID);my_genelistmygenes=my_genelist %>% lapply( function(x) {stringr::str_split(x,"/",simplify = TRUE) %>%as.vector() %>%unique()} )#准备画热图,加载seurat对象
load("/home/data/t040413/silicosis/spatial_transcriptomics/silicosis_ST_harmony_SCT_r0.5.rds")
{dim(d.all)DefaultAssay(d.all)="Spatial"#visium_slides=SplitObject(object = d.all,split.by = "stim")names(d.all);dim(d.all)d.all@meta.data %>%head()head(colnames(d.all))#1 给d.all 添加meta信息------adata_obs=read.csv("~/silicosis/spatial/adata_obs.csv")head(adata_obs)mymeta= paste0(d.all@meta.data$orig.ident,"_",colnames(d.all)) %>% gsub("-.*","",.) # %>% head()head(mymeta)tail(mymeta)#掉-及其之后内容adata_obs$col= adata_obs$spot_id %>% gsub("-.*","",.) # %>% head()head(adata_obs)rownames(adata_obs)=adata_obs$coladata_obs=adata_obs[mymeta,]head(adata_obs)identical(mymeta,adata_obs$col)d.all=AddMetaData(d.all,metadata = adata_obs)head(d.all@meta.data)}##构建画热图对象---
Idents(d.all)=d.all$clusters
a=AverageExpression(d.all,return.seurat = TRUE)
a$orig.ident=rownames(a@meta.data)
head(a@meta.data)
head(markers)rownames(a) %>%head()
head(mygenes)
table(mygenes %in% rownames(a))
DoHeatmap(a,draw.lines = FALSE, slot = 'scale.data', group.by = 'orig.ident',features = mygenes ) + ggplot2:: scale_color_discrete(name = "Identity", labels = unique(a$orig.ident) %>%sort() )##doheatmap做出来的图不好调整,换成heatmap自己调整p=DoHeatmap(a,draw.lines = FALSE, slot = 'scale.data', group.by = 'orig.ident',features = mygenes ) + ggplot2:: scale_color_discrete( labels = unique(a$orig.ident) %>%sort() ) #name = "Identity",p$data %>%head()##########这种方式容易出现bug,不建议------
if (F) {wide_data <- p$data %>% .[,-4] %>%tidyr:: pivot_wider(names_from = Cell, values_from = Expression)print(wide_data) mydata= wide_data %>%dplyr:: select(-Feature) %>%as.matrix()head(mydata)rownames(mydata)=wide_data$Featuremydata=mydata[,c("Bronchial zone", "Fibrogenic zone", "Interstitial zone", "Inflammatory zone","Vascular zone" )]p2=pheatmap:: pheatmap(mydata, fontsize_row = 2, clustering_method = "ward.D2",# annotation_col = wide_data$Feature,annotation_colors = c("Interstitial zone" = "red", "Bronchial zone" = "blue", "Fibrogenic zone" = "green", "Vascular zone" = "purple") ,cluster_cols = FALSE,column_order = c("Inflammatory zone", "Vascular zone" ,"Bronchial zone", "Fibrogenic zone" ))getwd()ggplot2::ggsave(filename = "~/silicosis/spatial/sp_cluster_rigions_after_harmony/heatmap_usingpheatmap.pdf",width = 8,height = 10,limitsize = FALSE,plot = p2)}##########建议如下方式画热图------
a$orig.ident=a@meta.data %>%rownames()
a@meta.data %>%head()
Idents(a)=a$orig.identa@assays$Spatial@scale.data %>%head()mydata=a@assays$Spatial@scale.data
mydata=mydata[rownames(mydata) %in% (mygenes %>%unlist() %>%unique()) ,]
mydata= mydata[,c( "Fibrogenic zone", "Inflammatory zone", "Bronchial zone","Interstitial zone","Vascular zone" )]
head(mydata)
p3=pheatmap:: pheatmap(mydata, fontsize_row = 2, clustering_method = "ward.D2",# annotation_col = wide_data$Feature,annotation_colors = c("Interstitial zone" = "red", "Bronchial zone" = "blue", "Fibrogenic zone" = "green", "Vascular zone" = "purple") ,cluster_cols = FALSE,column_order = c("Inflammatory zone", "Vascular zone" ,"Bronchial zone", "Fibrogenic zone" )
)getwd()
ggplot2::ggsave(filename = "~/silicosis/spatial/sp_cluster_rigions_after_harmony/heatmap_usingpheatmap2.pdf",width = 8,height = 10,limitsize = FALSE,plot = p3)#########单独画出炎症区和纤维化区---------
a@assays$Spatial@scale.data %>%head()mydata=a@assays$Spatial@scale.data
mygenes2= my_genelist[c('Inflammatory zone','Fibrogenic zone')] %>% unlist() %>% stringr::str_split("/",simplify = TRUE) mydata2=mydata[rownames(mydata) %in% ( mygenes2 %>%unlist() %>%unique()) ,]
mydata2= mydata2[,c( "Fibrogenic zone", "Inflammatory zone" )]
head(mydata2)p3=pheatmap:: pheatmap(mydata2, fontsize_row = 5, #scale = 'row',clustering_method = "ward.D2",# annotation_col = wide_data$Feature,annotation_colors = c("Interstitial zone" = "red", "Bronchial zone" = "blue", "Fibrogenic zone" = "green", "Vascular zone" = "purple") ,cluster_cols = FALSE,column_order = c("Inflammatory zone", "Vascular zone" ,"Bronchial zone", "Fibrogenic zone" )
)getwd()
ggplot2::ggsave(filename = "~/silicosis/spatial/sp_cluster_rigions_after_harmony/heatmap_usingpheatmap3.pdf",width = 4,height = 8,limitsize = FALSE,plot = p3)
.libPaths(c("/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2","/home/data/t040413/R/yll/usr/local/lib/R/site-library", "/usr/local/lib/R/library","/refdir/Rlib/"))library(Seurat)
library(ggplot2)
library(dplyr)
load('/home/data/t040413/silicosis/fibroblast_myofibroblast2/subset_data_fibroblast_myofibroblast2.rds')print(getwd())
dir.create("~/silicosis/fibroblast_myofibroblast2/slilica_fibroblast_enrichments")
setwd("~/silicosis/fibroblast_myofibroblast2/slilica_fibroblast_enrichments")
getwd()DimPlot(subset_data,label = TRUE)markers_foreach=FindAllMarkers(subset_data,only.pos = TRUE,densify = TRUE)
head(markers_foreach)all_degs=markers_foreach
markers=markers_foreach#install.packages("AnnotationDbi")
#BiocManager::install(version = '3.18')
#BiocManager::install('AnnotationDbi',version = '3.18')
{##########################---------------------批量富集分析findallmarkers-enrichment analysis==================================================#https://mp.weixin.qq.com/s/WyT-7yKB9YKkZjjyraZdPgdf=all_degs##筛选阈值确定:p<0.05,|log2FC|>1p_val_adj = 0.05# avg_log2FC = 0.1fc=seq(0.8,1.4,0.2)print(getwd())#setwd("../")for (avg_log2FC in fc) {# avg_log2FC=0.5dir.create(paste0(avg_log2FC,"/"))setwd(paste0(avg_log2FC,"/"))print(getwd())print(paste0("Start----",avg_log2FC))head(all_degs)#根据阈值添加上下调分组标签:df$direction <- case_when(df$avg_log2FC > avg_log2FC & df$p_val_adj < p_val_adj ~ "up",df$avg_log2FC < -avg_log2FC & df$p_val_adj < p_val_adj ~ "down",TRUE ~ 'none')head(df)df=df[df$direction!="none",]head(df)dim(df)df$mygroup=paste(df$group,df$direction,sep = "_")head(df)dim(df)##########################----------------------enrichment analysis=#https://mp.weixin.qq.com/s/WyT-7yKB9YKkZjjyraZdPg{library(clusterProfiler)library(org.Hs.eg.db) #人library(org.Mm.eg.db) #鼠library(ggplot2)# degs_for_nlung_vs_tlung$gene=rownames(degs_for_nlung_vs_tlung)head(markers)df=markers %>%dplyr::group_by(cluster)%>%filter(p_val_adj <0.05)sce.markers=dfhead(sce.markers)print(getwd())ids <- suppressWarnings(bitr(sce.markers$gene, 'SYMBOL', 'ENTREZID', 'org.Mm.eg.db'))head(ids)head(sce.markers)tail(sce.markers)dim(sce.markers)sce.markers=merge(sce.markers,ids,by.x='gene',by.y='SYMBOL')head(sce.markers)dim(sce.markers)sce.markers$group=sce.markers$clustersce.markers=sce.markers[sce.markers$group!="none",]dim(sce.markers)head(sce.markers)getwd()# sce.markers=openxlsx::read.xlsx("/home/data/t040413/silicosis/fibroblast_myofibroblast/group_enrichments/")#sce.markers$cluster=sce.markers$mygroupdim(sce.markers)head(sce.markers)gcSample=split(sce.markers$ENTREZID, sce.markers$cluster)library(clusterProfiler)gcSample # entrez id , compareCluster print("===========开始go= All ont===========")xx <- compareCluster(gcSample, fun="enrichGO",OrgDb="org.Mm.eg.db" , #'org.Hs.eg.db',pvalueCutoff=0.05) #organism="hsa",xx.BP <- compareCluster(gcSample, fun="enrichGO",OrgDb="org.Mm.eg.db" , #'org.Hs.eg.db',pvalueCutoff=0.05,readable=TRUE,ont="BP") #organism="hsa",p=clusterProfiler::dotplot(object = xx.BP,showCategory = 20,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-BP_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-BP_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)xx.CC <- compareCluster(gcSample, fun="enrichGO",OrgDb="org.Mm.eg.db" , #'org.Hs.eg.db',pvalueCutoff=0.05,readable=TRUE,ont="CC") #organism="hsa",p=clusterProfiler::dotplot(object = xx.CC,showCategory = 20,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-CC_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-CC_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)xx.MF <- compareCluster(gcSample, fun="enrichGO",OrgDb="org.Mm.eg.db" , #'org.Hs.eg.db',pvalueCutoff=0.05,readable=TRUE,ont="MF") #organism="hsa",p=clusterProfiler::dotplot(object = xx.MF,showCategory = 20,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-MF_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-MF_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)print(getwd()).libPaths()print("===========开始 kegg All ont============")gg<-clusterProfiler::compareCluster(gcSample,fun = "enrichKEGG", #readable=TRUE,keyType = 'kegg', #KEGG 富集organism='mmu',#"rno",pvalueCutoff = 0.05 #指定 p 值阈值(可指定 1 以输出全部)p=clusterProfiler::dotplot(object = xx,showCategory = 20,label_format =100) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-GO_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-GO_enrichment--3.pdf'),plot = p,width = 13,height = 40,limitsize = F)xxwrite.csv(xx,file = paste0(avg_log2FC,"compareCluster-GO_enrichment.csv"))p=clusterProfiler::dotplot(gg,showCategory = 20,label_format = 40) p4=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))p4print(paste("保存位置",getwd(),sep = " : "))ggsave(paste0(avg_log2FC,'_degs_compareCluster-KEGG_enrichment-2.pdf'),plot = p4,width = 13,height = 25,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-KEGG_enrichment-2.pdf'),plot = p4,width = 13,height = 25,limitsize = F)ggopenxlsx::write.xlsx(gg,file = paste0(avg_log2FC,"_compareCluster-KEGG_enrichment.xlsx"))getwd()openxlsx::write.xlsx(sce.markers,file = paste0(avg_log2FC,"_sce.markers_for_each_clusterfor_enrichment.xlsx"))##放大图片{getwd()#bpp=clusterProfiler::dotplot(object = xx.BP,showCategory = 100,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-BP_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-BP_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)print((getwd()))#ccp=clusterProfiler::dotplot(object = xx.CC,showCategory = 100,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-CC_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-CC_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)print((getwd()))#MFp=clusterProfiler::dotplot(object = xx.MF,showCategory = 100,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-MF_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-MF_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)print((getwd()))#KEGGp=clusterProfiler::dotplot(object = gg,showCategory = 100,label_format =60) p=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))pggsave(paste0(avg_log2FC,'_degs_compareCluster-KEGG_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)ggsave(paste0(avg_log2FC,'_degs_compareCluster-KEGG_enrichment-100terms-3.pdf'),plot = p,width = 13,height = 100,limitsize = F)print((getwd()))}# save(xx.BP,xx.CC,xx.MF, gg, file = "~/silicosis/spatial_transcriptomicsharmony_cluster_0.5res_gsea/xx.Rdata")xx.BP@compareClusterResult$oncology="BP"xx.CC@compareClusterResult$oncology="CC"xx.MF@compareClusterResult$oncology="MF"xx.all=do.call(rbind,list(xx.BP@compareClusterResult,xx.CC@compareClusterResult,xx.MF@compareClusterResult))head(xx.all) openxlsx::write.xlsx(xx.all,file = paste0(avg_log2FC,"_enrichments_all.xlsx")) # save(xx.BP,xx.CC,xx.MF,xx.all,sce.markers,merged_degs, gg, file = "./xx.Rdata")result <- tryCatch({save(xx.BP,xx.CC,xx.MF,xx.all,sce.markers,merged_degs, gg, file = paste0(avg_log2FC,"_xx.Rdata"))# 在这里添加你希望在没有报错时执行的代码print("没有报错")}, error = function(e) {print(getwd()) # 在报错时执行的代码# 在这里添加你希望在报错时执行的额外代码save(xx.BP,xx.CC,xx.MF,xx.all,sce.markers, gg, file = paste0(avg_log2FC,"_xx_all.Rdata"))})# print(result)print(getwd())if (F) { #大鼠print("===========开始go============")xx <-clusterProfiler::compareCluster(gcSample, fun="enrichGO",OrgDb="org.Rn.eg.db",readable=TRUE,ont = 'ALL', #GO Ontology,可选 BP、MF、CC,也可以指定 ALL 同时计算 3 者pvalueCutoff=0.05) #organism="hsa", #'org.Hs.eg.db',print("===========开始 kegg============")gg<-clusterProfiler::compareCluster(gcSample,fun = "enrichKEGG",keyType = 'kegg', #KEGG 富集organism="rno",pvalueCutoff = 0.05 #指定 p 值阈值(可指定 1 以输出全部)p=dotplot(xx) p2=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))p2ggsave('degs_compareCluster-GO_enrichment-2.pdf',plot = p2,width = 6,height = 20,limitsize = F)xxopenxlsx::write.xlsx(xx,file = "compareCluster-GO_enrichment.xlsx")#p=dotplot(gg) p4=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5))p4print(paste("保存位置",getwd(),sep = " : "))ggsave('degs_compareCluster-KEGG_enrichment-2.pdf',plot = p4,width = 6,height = 12,limitsize = F)ggopenxlsx::write.xlsx(gg,file = "compareCluster-KEGG_enrichment.xlsx") } setwd("../")}}}print(getwd())
#setwd('../')load("~/silicosis/fibroblast_myofibroblast2/slilica_fibroblast_enrichments/1/1_xx_all.Rdata")# remove duplicate rows based on Description 并且保留其他所有变量
xx.all<- distinct(xx.all, Description,.keep_all = TRUE)enrichmets=xx.all#2.2挑选term---selected_clusterenrich=enrichmets[grepl(pattern = "lung alveolus developmen|extracellular matrix organization|extracellular structure organization|response to toxic substance|blood vessel endothelial cell migrationregulation of blood vessel endothelial cell migration|lung development|respiratory tube development|detoxification|cellular response to toxic substance|endothelial cell proliferation|cellular response to chemical stress|Wnt|infla|fibrob|myeloid",x = enrichmets$Description) &stringr::str_detect(pattern = "muscle|fibroblast migration|positive regulation of endothelial cell proliferation|inflammatory response to wounding|wound healing involved in inflammatory response|fibronectin binding|fibronectin binding|inflammatory response to wounding|fibronectin binding", negate = TRUE, string = enrichmets$Description),]head(selected_clusterenrich) distinct(selected_clusterenrich)# remove duplicate rows based on Description 并且保留其他所有变量
distinct_df <- distinct(selected_clusterenrich, Description,.keep_all = TRUE)ggplot( distinct(selected_clusterenrich,Description,.keep_all=TRUE) %>% # dplyr::mutate(Cluster = factor(Cluster, levels = unique(.$Cluster))) %>%dplyr::mutate(Description = factor(Description, levels = unique(.$Description))) %>%# dplyr::group_by(Cluster) %>%dplyr::filter(stringr::str_detect(pattern = "muscle", negate = TRUE,Description)) %>%add_count() %>%dplyr::arrange(dplyr::desc(n),dplyr::desc(Description)) %>%mutate(Description =forcats:: fct_inorder(Description)), #fibri|matrix|collaaes(Cluster, y = Description)) + #stringr:: str_wrapscale_y_discrete(labels = function(x) stringr::str_wrap(x, width = 58)) + #调整terms长度 字符太长geom_point(aes(fill=p.adjust, size=Count), shape=21)+theme_bw()+theme(axis.text.x=element_text(angle=90,hjust = 1,vjust=0.5),axis.text.y=element_text(size = 12),axis.text = element_text(color = 'black', size = 12))+scale_fill_gradient(low="red",high="blue")+labs(x=NULL,y=NULL)
# coord_flip()
print(getwd())

![给定n个字符串s[1...n], 求有多少个数对(i, j), 满足i < j 且 s[i] + s[j] == s[j] + s[i]?](https://img-blog.csdnimg.cn/direct/85d295adffbd4ef8897e17df713e7440.jpeg)