MedSAM 项目排坑记录
- 任务
- 排坑过程
- 配置python环境
- 测试
- 构建docker
- 模型训练
- 数据预处理
- 单GPU训练
- 最后推理
任务
做一个课程大作业,需要进行CVPR2024年医疗影像分割赛题的打榜(CVPR 2024: SEGMENT ANYTHING IN MEDICAL IMAGES ON LAPTOP)。看到官方给了一个baseline,也就是这个MedSAM,决定先在我本地把这个baseline跑出来试试效果。
排坑过程
配置python环境
创建conda环境:
conda create -n MEDSAM python=3.10 -y
conda activate MEDSAM
安装pytorch:
pip3 install torch
中间竟然报错我磁盘空间没有了。害,存的数据集太多了。清理了一点,还有点暂时舍不得清理。主要是有一个去年上智能机器人课时候,跑SLAM算法大作业时下载的KITTI数据集和其他几个数据集,占了400多G空间,还有之前下载的HM3D数据集100多G。打算这两天把那个项目代码再跑一遍,上传到平台上,然后把本地的删除。以后可能还要下载很多数据集,打算下血本买个西数的p40 2T硬盘。正好我电脑有雷电4接口,能发挥硬盘宣称的2000M速度。
克隆项目:
git clone -b LiteMedSAM https://github.com/bowang-lab/MedSAM/
其中-b是指定分支branch。
安装项目包:
pip3 install -e .
据这个博客参考博客所说,这个命令就是将当前项目以软链接并且可修改的形式安装到当前python环境中,具体来说就是执行setup.py脚本。
然后经典报错:
ERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.
rosdep 0.22.2 requires catkin-pkg>=0.4.0, which is not installed.
rosdistro 0.9.0 requires catkin-pkg, which is not installed.
rosinstall 0.7.8 requires catkin-pkg, which is not installed.
rosinstall-generator 0.1.23 requires catkin-pkg>=0.1.28, which is not installed.
rospkg 1.5.0 requires catkin-pkg, which is not installed.
rospkg 1.5.0 requires distro>=1.4.0; python_version >= "3.8", which is not installed.
安装就好:
pip3 install empy==3.3.4 rospkg pyyaml catkin_pkg
测试
先要下载模型参数。打开work_dir/LiteMedSAM这个目录,发现里面有个readme,点开其中的链接,从那个链接的谷歌云盘上下载下来,放到这个目录就行。
接着下载示例数据,从那个here点进去,下载三个谷歌云盘文件。然后把他们放到test_demo这个文件夹下面。
最后运行推理脚本:
python CVPR24_LiteMedSAM_infer.py -i test_demo/imgs/ -o test_demo/segs
结果为:
感觉跟没跑一样。
看代码:
if __name__ == '__main__':img_npz_files = sorted(glob(join(data_root, '*.npz'), recursive=True))print(img_npz_files)efficiency = OrderedDict()efficiency['case'] = []efficiency['time'] = []for img_npz_file in tqdm(img_npz_files):print('in')start_time = time()if basename(img_npz_file).startswith('3D'):MedSAM_infer_npz_3D(img_npz_file)else:MedSAM_infer_npz_2D(img_npz_file)end_time = time()efficiency['case'].append(basename(img_npz_file))efficiency['time'].append(end_time - start_time)current_time = datetime.now().strftime("%Y-%m-%d %H:%M:%S")print(current_time, 'file name:', basename(img_npz_file), 'time cost:', np.round(end_time - start_time, 4))efficiency_df = pd.DataFrame(efficiency)efficiency_df.to_csv(join(pred_save_dir, 'efficiency.csv'), index=False)
我打印出img_npz_files,发现是空列表。说明没读到文件啊。然后发现我是呆比:数据都没解压缩。解压后再运行:
python /home/lcy-magic/MedSAM/MedSAM/CVPR24_LiteMedSAM_infer.py -i /home/lcy-magic/MedSAM/MedSAM/test_demo/imgs/ -o test_demo/segs
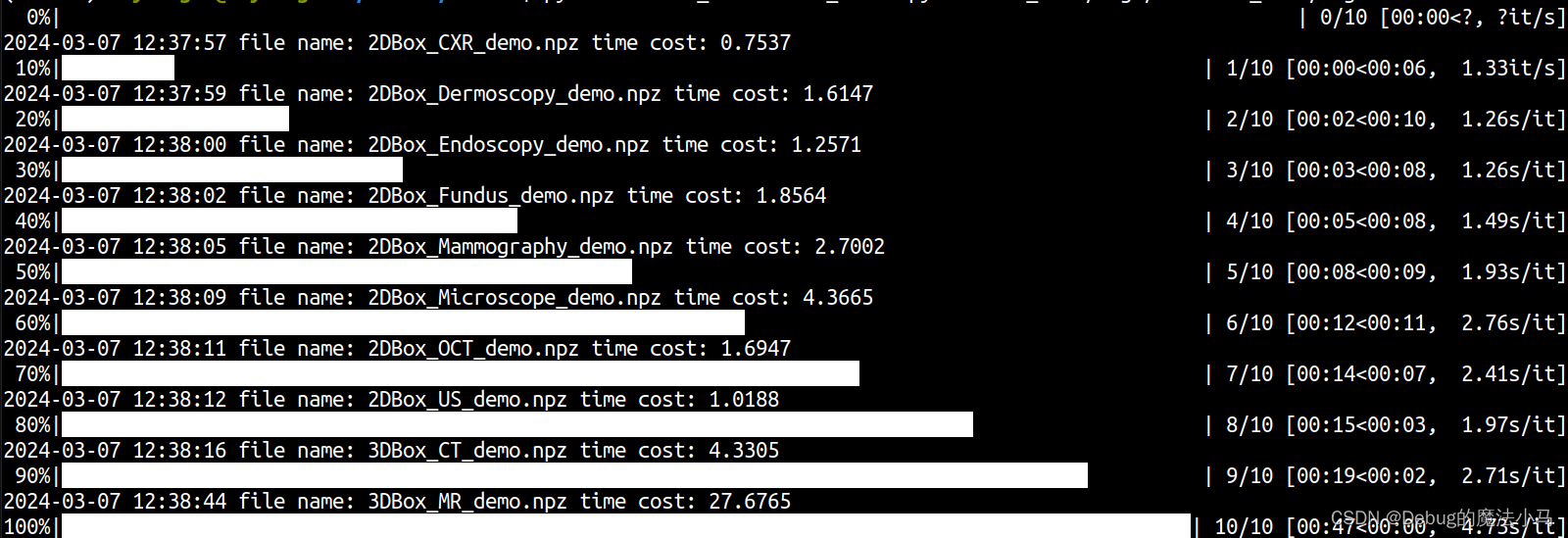
这回就好了:
构建docker
在MedSAM目录下运行:
docker build -f Dockerfile -t litemedsam .
会用很长时间。
测试docker。先给可执行权限:
sudo chmod -R 777 ./*
然后运行:
docker container run -m 8G --name litemedsam --rm -v $PWD/test_demo/imgs/:/workspace/inputs/ -v $PWD/test_demo/litemedsam-seg/:/workspace/outputs/ litemedsam:latest /bin/bash -c "sh predict.sh"
其中PWD这个环境变量是当前路径,所以也要在在MedSAM目录下运行。
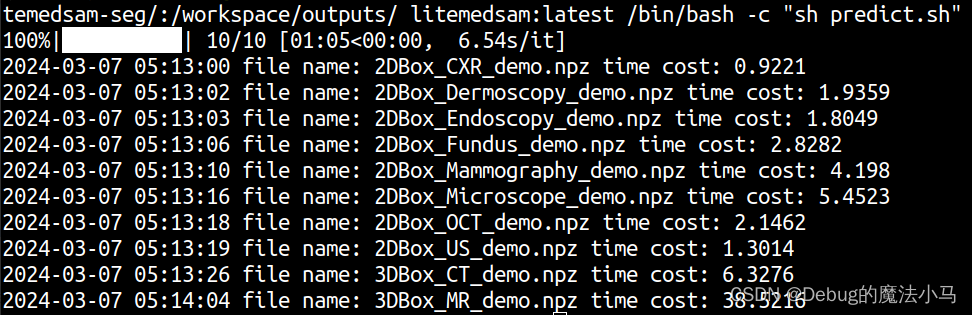
成功:
保存docker为一个压缩文件:
docker save litemedsam | gzip -c > litemedsam.tar.gz
时间也比较漫长。成功:

最后运行:
python /home/lcy-magic/MedSAM/MedSAM/evaluation/compute_metrics.py -s test_demo/litemedsam-seg -g test_demo/gts -csv_dir ./metrics.csv
没问题
模型训练
数据预处理
首先模型参数咱已经有了,应该不用再下载了。但要下载数据。在他的demo dataset链接里,下载这个压缩包:
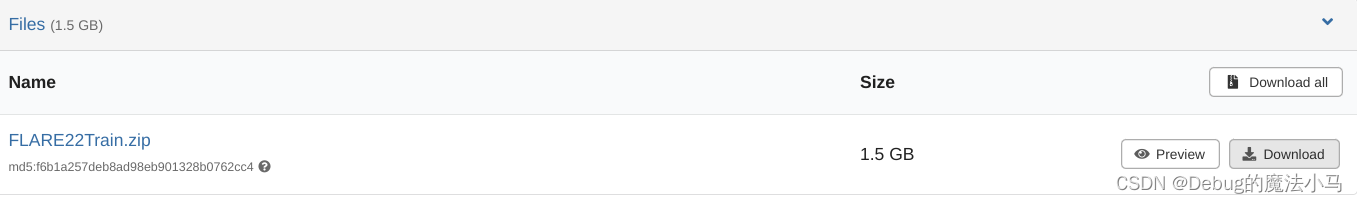
然后再MedSAM文件夹下新建data文件夹,把这个压缩包放过去解压缩。
数据处理那个指令太长了,写个shell脚本吧:
touch pre_CT_MR.sh
chmod 777 pre_CT_MR.sh
gedit pre_CT_MR.sh
写入以下语句(参考:参考博客):
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python pre_CT_MR.py \-img_path data/FLARE22Train/images \-img_name_suffix _0000.nii.gz \-gt_path data/FLARE22Train/labels \-gt_name_suffix .nii.gz \-output_path data \-num_workers 4 \-prefix CT_Abd_ \-modality CT \-anatomy Abd \-window_level 40 \-window_width 400 \--save_nii
执行脚本:
./pre_CT_MR.sh
报错:
./pre-processing.sh: 行 3: activate: 没有那个文件或目录
Traceback (most recent call last):File "/home/lcy-magic/MedSAM/MedSAM/pre_CT_MR.py", line 12, in <module>import cc3d
ModuleNotFoundError: No module named 'cc3d'
./pre-processing.sh: 行 7: -img_name_suffix:未找到命令
./pre-processing.sh: 行 8: -gt_path:未找到命令
./pre-processing.sh: 行 9: -gt_name_suffix:未找到命令
./pre-processing.sh: 行 10: -output_path:未找到命令
./pre-processing.sh: 行 11: -num_workers:未找到命令
./pre-processing.sh: 行 12: -prefix:未找到命令
./pre-processing.sh: 行 13: -modality:未找到命令
./pre-processing.sh: 行 14: -anatomy:未找到命令
./pre-processing.sh: 行 15: -window_level:未找到命令
./pre-processing.sh: 行 16: -window_width:未找到命令
./pre-processing.sh: 行 17: --save_nii:未找到命令
那就安装cc3d:
pip3 install connected-components-3d
再运行,又报错:
usage: pre_CT_MR.py [-h] [-modality MODALITY] [-anatomy ANATOMY] [-img_name_suffix IMG_NAME_SUFFIX] [-gt_name_suffix GT_NAME_SUFFIX] [-img_path IMG_PATH][-gt_path GT_PATH] [-output_path OUTPUT_PATH] [-num_workers NUM_WORKERS] [-window_level WINDOW_LEVEL] [-window_width WINDOW_WIDTH][--save_nii]
pre_CT_MR.py: error: unrecognized arguments: -prefix CT_Abd_
查看python脚本可以发现,其他参数都是读取的,CT_Abd_参数是根据其他参数拼接得到的。而且结果和命令行给出的一致。那就不再需要命令行指定了。
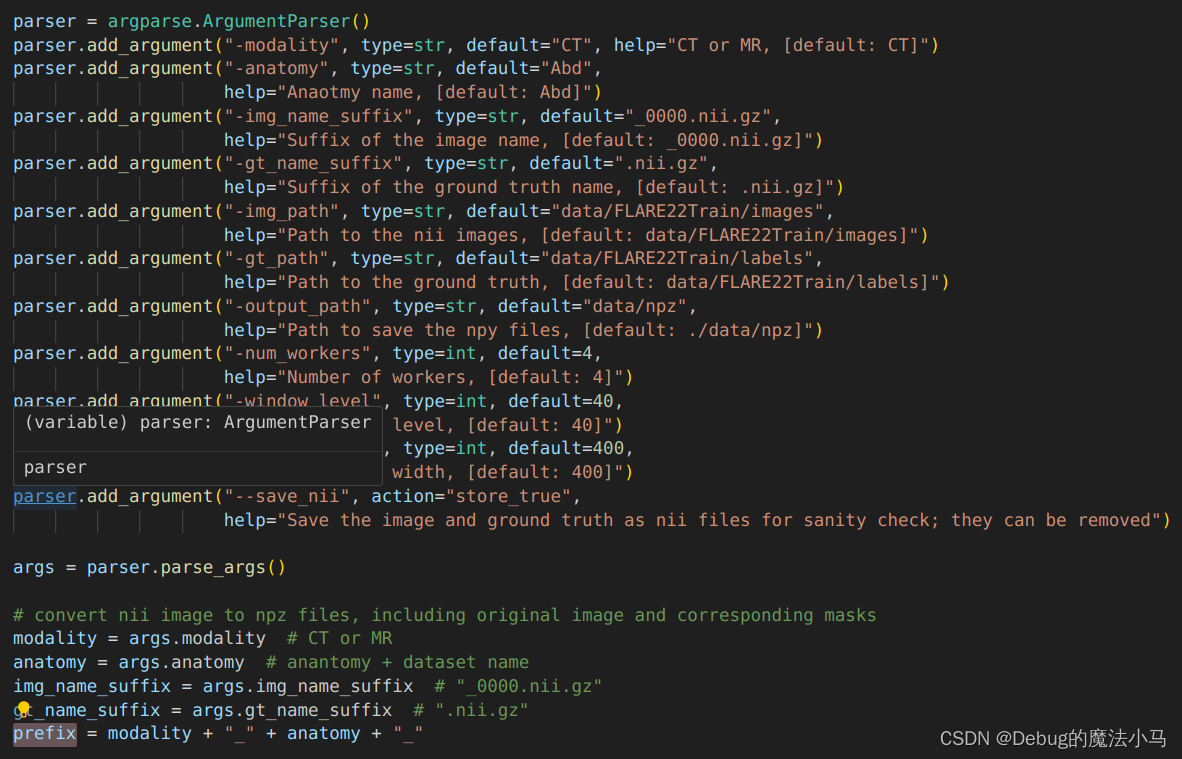
于是我简单粗暴地把这一行删掉了,再运行没问题:

接下来做格式转换,照样改成shell脚本:
touch npz_to_npy.sh
chmod 777 npz_to_npy.sh
gedit npz_to_npy.sh
写入:
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python npz_to_npy.py \-npz_dir data/MedSAM_train \-npy_dir data/npy \-num_workers 4
运行:
./npz_to_npy.sh
结果:

感觉是文件路径不对,看了下文件。果然,训练数据还在下一级目录。应该把脚本改成:
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python npz_to_npy.py \-npz_dir data/MedSAM_train/CT_Abd \-npy_dir data/npy \-num_workers 4
再运行就没问题了:
Converting npz to npy: 100%|███████████████████████████████████████████████████████████████████████████████████████████████████| 40/40 [00:05<00:00, 7.55it/s]单GPU训练
同样,构建脚本:
touch train_one_gpu.sh
chmod 777 train_one_gpu.sh
gedit train_one_gpu.sh
写入:
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python train_one_gpu.py \-data_root data/MedSAM_train/CT_Abd \-pretrained_checkpoint lite_medsam.pth \-work_dir work_dir \-num_workers 4 \-batch_size 4 \-num_epochs 10# ~/anaconda3/envs/MEDSAM/bin/python train_one_gpu.py \
# -data_root data/MedSAM_train/CT_Abd \
# -resume work_dir/medsam_lite_latest.pth \
# -work_dir work_dir \
# -num_workers 4 \
# -batch_size 4 \
# -num_epochs 10
如果从lite_medsam开始微调就注释下面的,如果从上次中断的训练开始微调就注释上面的。
运行:
./train_one_gpu.sh
报错:
Pretained weights lite_medsam.pth not found, training from scratch
MedSAM Lite size: 9791300
Traceback (most recent call last):File "/home/lcy-magic/MedSAM/MedSAM/train_one_gpu.py", line 408, in <module>train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True, num_workers=num_workers, pin_memory=True)File "/home/lcy-magic/anaconda3/envs/MEDSAM/lib/python3.10/site-packages/torch/utils/data/dataloader.py", line 350, in __init__sampler = RandomSampler(dataset, generator=generator) # type: ignore[arg-type]File "/home/lcy-magic/anaconda3/envs/MEDSAM/lib/python3.10/site-packages/torch/utils/data/sampler.py", line 143, in __init__raise ValueError(f"num_samples should be a positive integer value, but got num_samples={self.num_samples}")
ValueError: num_samples should be a positive integer value, but got num_samples=0
哦,他这个readme又写错了。现在训练用的是npy数据不是npz的。所以脚本应该改成:
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python train_one_gpu.py \-data_root data/npy \-pretrained_checkpoint lite_medsam.pth \-work_dir work_dir \-num_workers 4 \-batch_size 4 \-num_epochs 10# ~/anaconda3/envs/MEDSAM/bin/python train_one_gpu.py \
# -data_root data/npy \
# -resume work_dir/medsam_lite_latest.pth \
# -work_dir work_dir \
# -num_workers 4 \
# -batch_size 4 \
# -num_epochs 10
再次运行,报错:
torch.cuda.OutOfMemoryError: CUDA out of memory. Tried to allocate 36.00 MiB. GPU 0 has a total capacity of 7.79 GiB of which 47.56 MiB is free. Including non-PyTorch memory, this process has 6.72 GiB memory in use. Of the allocated memory 6.42 GiB is allocated by PyTorch, and 123.97 MiB is reserved by PyTorch but unallocated. If reserved but unallocated memory is large try setting PYTORCH_CUDA_ALLOC_CONF=expandable_segments:True to avoid fragmentation. See documentation for Memory Management (https://pytorch.org/docs/stable/notes/cuda.html#environment-variables)
又没显存了。。。而且batch_size已经是4了,很小了。我试了下,只有改成1或者2我这里才不报错。这两天有空还是尝试把核显跟独显的功能分开吧,多省点显存。
但也会有新报错:
Traceback (most recent call last):File "/home/lcy-magic/MedSAM/MedSAM/train_one_gpu.py", line 443, in <module>loss.backward()File "/home/lcy-magic/anaconda3/envs/MEDSAM/lib/python3.10/site-packages/torch/_tensor.py", line 522, in backwardtorch.autograd.backward(File "/home/lcy-magic/anaconda3/envs/MEDSAM/lib/python3.10/site-packages/torch/autograd/__init__.py", line 266, in backwardVariable._execution_engine.run_backward( # Calls into the C++ engine to run the backward pass
RuntimeError: GET was unable to find an engine to execute this computation
也就是找不到反向传播的工具了。
什么原因呐?好像是显卡驱动问题,但有人说他重装一下torch就好了,我也打算这样先试一下参考issue:
pip uninstall torch
pip3 install torch torchvision
结果还是不行。我想我的torch是2.2.1的,这个项目要求是2.1.2的。我回退一下版本试试?
pip3 install torch==2.1.2 torchvision
再运行还是同样的错误。那应该就是我的驱动版本问题了。
检查驱动版本:
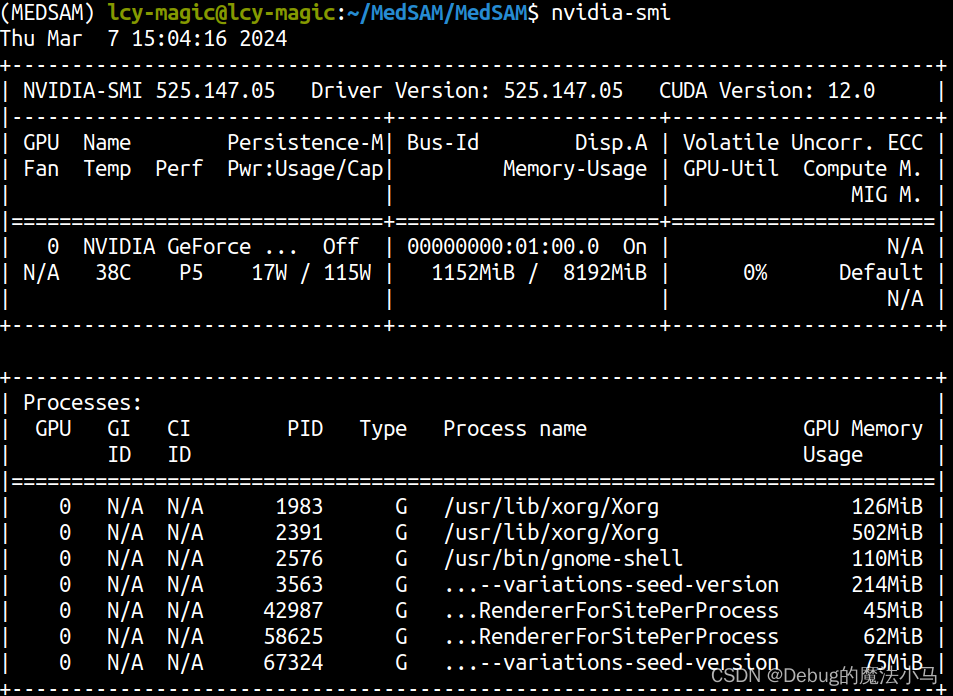
这个时候我发现我忽略了前面一大堆的报错信息:
Error: /usr/local/cuda/lib64/libcudnn_cnn_train.so.8: undefined symbol: _ZTIN10cask_cudnn14BaseKernelInfoE, version libcudnn_cnn_infer.so.8
Could not load library libcudnn_cnn_train.so.8. Error: /usr/local/cuda/lib64/libcudnn_cnn_train.so.8: undefined symbol: _ZTIN10cask_cudnn14BaseKernelInfoE, version libcudnn_cnn_infer.so.8
搜了一下这个,我就明白了。根据参考博客所说,应该是之前有个系统的cudnn,pip安装torch又有一个cudnn。指向错了,指到系统的了。反正我一般都用pip安装torch,不源码安装,我就把系统的cundnn删掉就好。我没有删,先按照博主所说,把bashrc那一行注释掉了:
#export CUDA_HOME=/usr/local/cuda
#export LD_LIBRARY_PATH=${CUDA_HOME}/lib64:${LD_LIBRARY_PATH}
#export PATH=${CUDA_HOME}/bin:${PATH}
也source了,但还是同样报错。我一怒之下,还是删了吧:
cd /usr/local/cuda/lib64
sudo rm -f libcudnn*
cd /usr/local/cuda/include
sudo rm -f cudnn*
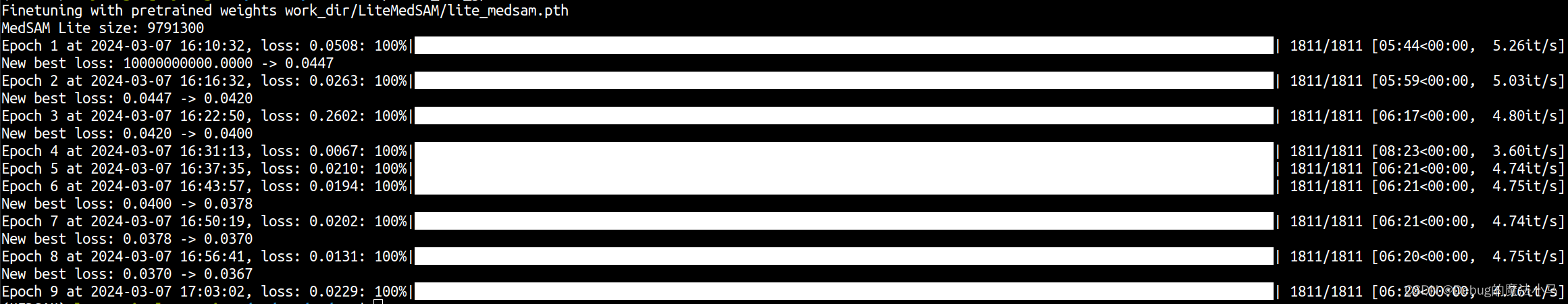
再回去运行bash脚本,就没问题了:

慢慢等结果了。遛个弯回来,发现到epoch3时候报错终止了。报错很长,主要是这部分:
RuntimeError: main thread is not in main loop
Tcl_AsyncDelete: async handler deleted by the wrong thread
参照参考博客进行解决。修改为:
# from matplotlib import pyplot as plt
import matplotlib
matplotlib.use('Agg')
from matplotlib import pyplot as plt
还发现,那个sh还有个地址不对,应该把模型参数地址改为:
-pretrained_checkpoint work_dir/LiteMedSAM/lite_medsam.pth \
这次训练完成:
最后推理
老样子:
touch train_one_gpu.sh
chmod 777 train_one_gpu.sh
gedit train_one_gpu.sh
写入:
#!/bin/bash~/anaconda3/envs/MEDSAM/bin/python inference_3D.py \-data_root data/MedSAM_test/CT_Abd \-pred_save_dir ./preds/CT_Abd \-medsam_lite_checkpoint_path work_dir/medsam_lite_latest.pth \-num_workers 4 \--save_overlay \-png_save_dir ./preds/CT_Abd_overlay \--overwrite
运行:
./inference_3D.sh
又报错报了一堆,提取最主要的:
Traceback (most recent call last):File "/home/lcy-magic/MedSAM/MedSAM/inference_3D.py", line 319, in <module>medsam_lite_model.load_state_dict(medsam_lite_checkpoint)File "/home/lcy-magic/anaconda3/envs/MEDSAM/lib/python3.10/site-packages/torch/nn/modules/module.py", line 2153, in load_state_dictraise RuntimeError('Error(s) in loading state_dict for {}:\n\t{}'.format(
RuntimeError: Error(s) in loading state_dict for MedSAM_Lite:Missing key(s) in state_dict:
和:
Unexpected key(s) in state_dict: "model", "epoch", "optimizer", "loss", "best_loss".
有missing key还有unexpected key,感觉像是整个文件搞错了,不是某一句的问题。
报错发生在medsam_lite_model.load_state_dict(medsam_lite_checkpoint)查看代码可以知道medsam_lite_checkpoint来自于bash脚本提供的-medsam_lite_checkpoint_path work_dir/medsam_lite_latest.pth \。
查看发现缺失是有这个文件的。不知道具体错哪了,换成medsam_lite_best.pth也不对,换成work_dir/LiteMedSAM/lite_medsam.pth \就好了:
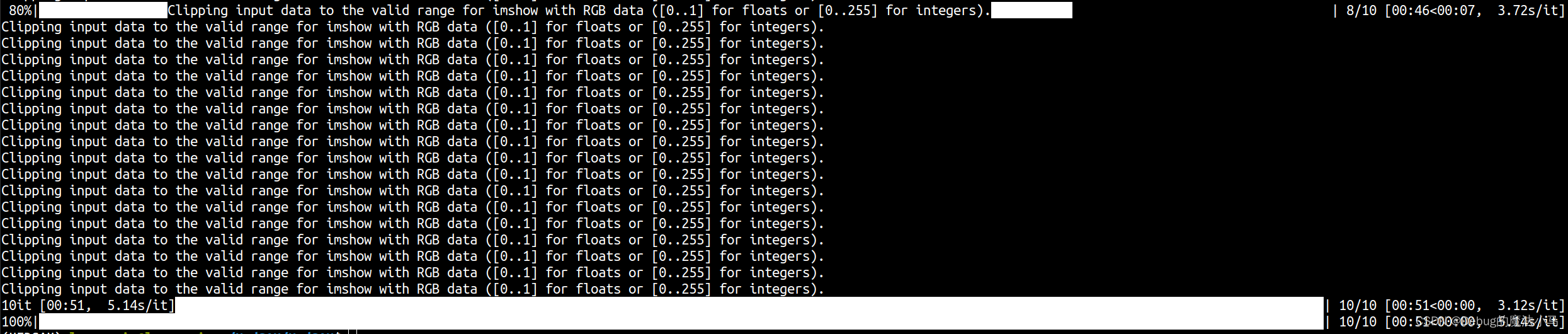
但这个应该是最初下载下来的,不是我训练的。在加载模型后打印一下看看:
medsam_lite_checkpoint = torch.load(medsam_lite_checkpoint_path, map_location='cpu')
print(medsam_lite_checkpoint.keys())
对于work_dir/medsam_lite_latest.pth \:
dict_keys(['model', 'epoch', 'optimizer', 'loss', 'best_loss'])
对于work_dir/LiteMedSAM/lite_medsam.pth \:
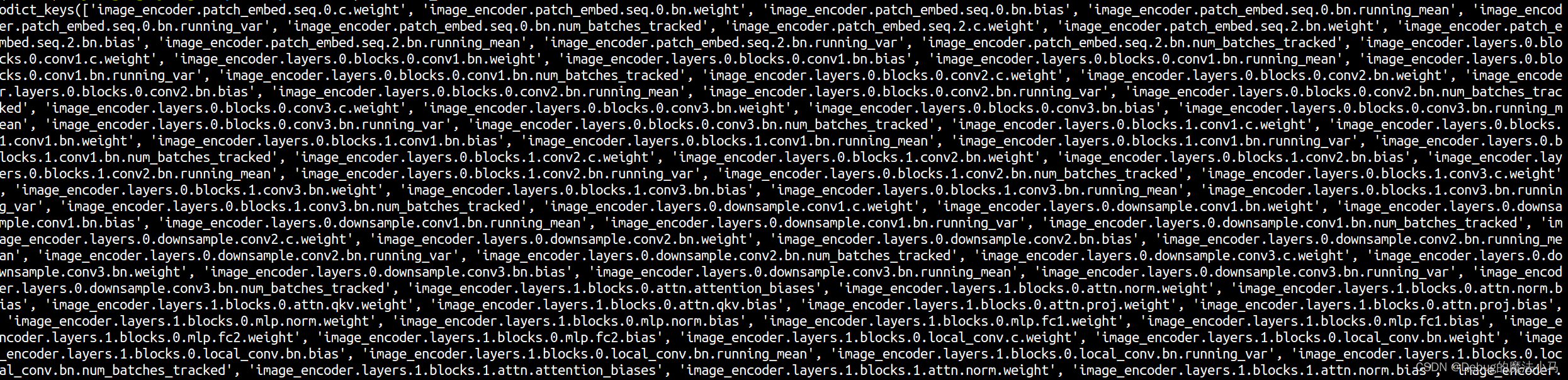
看到这个博客参考博客懂了,因为训练用的GPU,测试用的CPU,所以出的问题。改为:
medsam_lite_model.load_state_dict(medsam_lite_checkpoint,False)
就正常了。但这可能不是正确的解决办法。
测试一下:
这是之前的结果图:
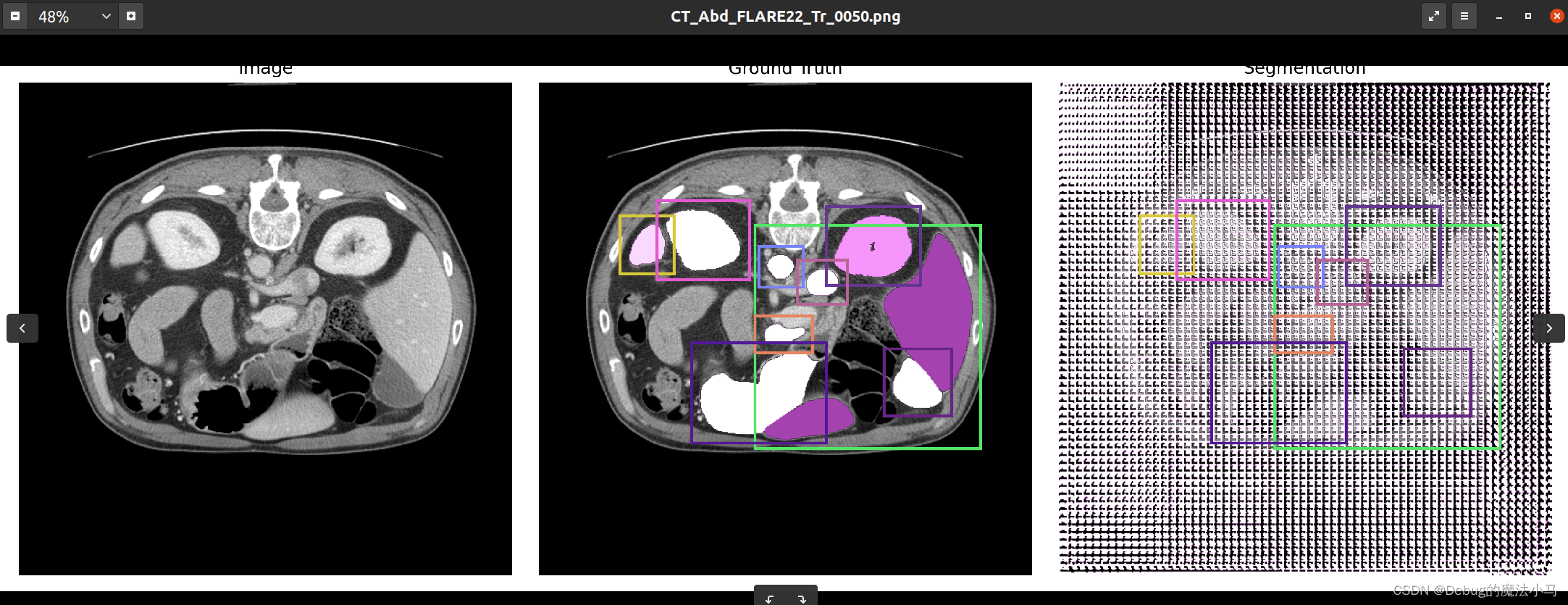
我把save_overlay参数设为True,运行后:
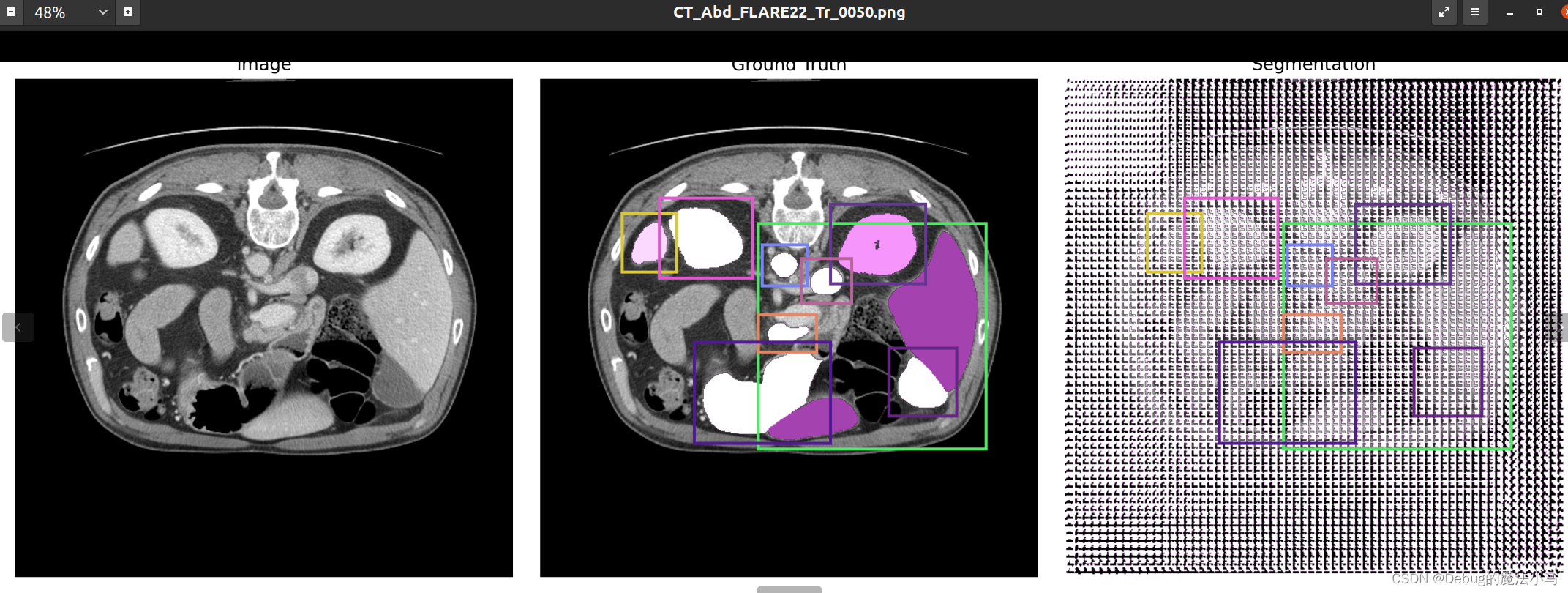
似乎没啥问题。暂时先这样了。