1、Yolov5的网络结构
- Yolov5中使用的Coco数据集输入图片的尺寸为640*640,但是训练过程的输入尺寸并不唯一,Yolov5可以采用Mosaic增强技术把4张图片的部分组成了一张尺寸一定的输入图片。如果需要使用预训练权重,最好将输入图片尺寸调整到与作者相同的尺寸,输入图片尺寸必须是32的倍数,这与anchor检测的阶段有关。
Yolov5s网络结构示意图:
- 当输入尺寸为640*640时,会得到3个不同尺度的输出:80x80(640/8)、40x40(640/16)、20x20(640/32)。
anchors:- [10, 13, 16, 30, 33, 23] # P3/8- [30, 61, 62, 45, 59, 119] # P4/16- [116, 90, 156, 198, 373, 326] # P5/32- anchors参数共有三行,每行6个数值,代表应用不同的特征图:
- 第一行是在最大的特征图上的锚框,80x80代表浅层的特征图(P3),包含较多的低层级信息,适合用于检测小目标,所以这一特征图所用的anchor尺度较小;
- 第二行是在中间的特征图上的锚框,40x40代表中间的特征图(P4),介于浅层和深层这两个尺度之间的anchor用来检测中等大小的目标;
- 第三行是在最小的特征图上的锚框,20x20代表深层的特征图(P5),包含更多高层级的信息,如轮廓、结构等信息,适合用于大目标的检测,所以这一特征图所用的anchor尺度较大。
待验证注释:
查阅其他博主博客发现,Yolov5也可以不预设anchor,直接写个3,此时yolov5就会自动按照训练集聚类anchor:
# Parameters nc: 4 # number of classes depth_multiple: 0.33 # model depth multiple width_multiple: 0.50 # layer channel multiple anchors: 3
在目标检测任务中,一般希望在大的特征图上去检测小目标,因为大特征图含有更多小目标信息,因此大特征图上的anchor数值通常设置为小数值,而小特征图上数值设置为大数值检测大的目标,yolov5之所以能高效快速地检测跨尺度目标,这种对不同特征图使用不同尺度的anchor的思想功不可没。
2、自适应锚框计算
- Yolov5 中并不是只使用默认锚定框,在开始训练之前会对数据集中标注信息进行核查,计算此数据集标注信息针对默认锚定框的最佳召回率。当最佳召回率大于或等于0.98,则不需要更新锚定框;如果最佳召回率小于0.98,则需要重新计算符合此数据集的锚定框。
- 核查锚定框是否适合要求的函数在 ./utils/autoanchor.py 文件中:
# YOLOv5 🚀 by Ultralytics, AGPL-3.0 license
"""AutoAnchor utils."""import randomimport numpy as np
import torch
import yaml
from tqdm import tqdmfrom utils import TryExcept
from utils.general import LOGGER, TQDM_BAR_FORMAT, colorstrPREFIX = colorstr("AutoAnchor: ")def check_anchor_order(m):"""Checks and corrects anchor order against stride in YOLOv5 Detect() module if necessary."""a = m.anchors.prod(-1).mean(-1).view(-1) # mean anchor area per output layerda = a[-1] - a[0] # delta ads = m.stride[-1] - m.stride[0] # delta sif da and (da.sign() != ds.sign()): # same orderLOGGER.info(f"{PREFIX}Reversing anchor order")m.anchors[:] = m.anchors.flip(0)@TryExcept(f"{PREFIX}ERROR")
def check_anchors(dataset, model, thr=4.0, imgsz=640):"""Evaluates anchor fit to dataset and adjusts if necessary, supporting customizable threshold and image size."""m = model.module.model[-1] if hasattr(model, "module") else model.model[-1] # Detect()shapes = imgsz * dataset.shapes / dataset.shapes.max(1, keepdims=True)scale = np.random.uniform(0.9, 1.1, size=(shapes.shape[0], 1)) # augment scalewh = torch.tensor(np.concatenate([l[:, 3:5] * s for s, l in zip(shapes * scale, dataset.labels)])).float() # whdef metric(k): # compute metricr = wh[:, None] / k[None]x = torch.min(r, 1 / r).min(2)[0] # ratio metricbest = x.max(1)[0] # best_xaat = (x > 1 / thr).float().sum(1).mean() # anchors above thresholdbpr = (best > 1 / thr).float().mean() # best possible recallreturn bpr, aatstride = m.stride.to(m.anchors.device).view(-1, 1, 1) # model stridesanchors = m.anchors.clone() * stride # current anchorsbpr, aat = metric(anchors.cpu().view(-1, 2))s = f"\n{PREFIX}{aat:.2f} anchors/target, {bpr:.3f} Best Possible Recall (BPR). "if bpr > 0.98: # threshold to recomputeLOGGER.info(f"{s}Current anchors are a good fit to dataset ✅")else:LOGGER.info(f"{s}Anchors are a poor fit to dataset ⚠️, attempting to improve...")na = m.anchors.numel() // 2 # number of anchorsanchors = kmean_anchors(dataset, n=na, img_size=imgsz, thr=thr, gen=1000, verbose=False)new_bpr = metric(anchors)[0]if new_bpr > bpr: # replace anchorsanchors = torch.tensor(anchors, device=m.anchors.device).type_as(m.anchors)m.anchors[:] = anchors.clone().view_as(m.anchors)check_anchor_order(m) # must be in pixel-space (not grid-space)m.anchors /= strides = f"{PREFIX}Done ✅ (optional: update model *.yaml to use these anchors in the future)"else:s = f"{PREFIX}Done ⚠️ (original anchors better than new anchors, proceeding with original anchors)"LOGGER.info(s)def kmean_anchors(dataset="./data/coco128.yaml", n=9, img_size=640, thr=4.0, gen=1000, verbose=True):"""Creates kmeans-evolved anchors from training dataset.Arguments:dataset: path to data.yaml, or a loaded datasetn: number of anchorsimg_size: image size used for trainingthr: anchor-label wh ratio threshold hyperparameter hyp['anchor_t'] used for training, default=4.0gen: generations to evolve anchors using genetic algorithmverbose: print all resultsReturn:k: kmeans evolved anchorsUsage:from utils.autoanchor import *; _ = kmean_anchors()"""from scipy.cluster.vq import kmeansnpr = np.randomthr = 1 / thrdef metric(k, wh): # compute metricsr = wh[:, None] / k[None]x = torch.min(r, 1 / r).min(2)[0] # ratio metric# x = wh_iou(wh, torch.tensor(k)) # iou metricreturn x, x.max(1)[0] # x, best_xdef anchor_fitness(k): # mutation fitness_, best = metric(torch.tensor(k, dtype=torch.float32), wh)return (best * (best > thr).float()).mean() # fitnessdef print_results(k, verbose=True):k = k[np.argsort(k.prod(1))] # sort small to largex, best = metric(k, wh0)bpr, aat = (best > thr).float().mean(), (x > thr).float().mean() * n # best possible recall, anch > thrs = (f"{PREFIX}thr={thr:.2f}: {bpr:.4f} best possible recall, {aat:.2f} anchors past thr\n"f"{PREFIX}n={n}, img_size={img_size}, metric_all={x.mean():.3f}/{best.mean():.3f}-mean/best, "f"past_thr={x[x > thr].mean():.3f}-mean: ")for x in k:s += "%i,%i, " % (round(x[0]), round(x[1]))if verbose:LOGGER.info(s[:-2])return kif isinstance(dataset, str): # *.yaml filewith open(dataset, errors="ignore") as f:data_dict = yaml.safe_load(f) # model dictfrom utils.dataloaders import LoadImagesAndLabelsdataset = LoadImagesAndLabels(data_dict["train"], augment=True, rect=True)# Get label whshapes = img_size * dataset.shapes / dataset.shapes.max(1, keepdims=True)wh0 = np.concatenate([l[:, 3:5] * s for s, l in zip(shapes, dataset.labels)]) # wh# Filteri = (wh0 < 3.0).any(1).sum()if i:LOGGER.info(f"{PREFIX}WARNING ⚠️ Extremely small objects found: {i} of {len(wh0)} labels are <3 pixels in size")wh = wh0[(wh0 >= 2.0).any(1)].astype(np.float32) # filter > 2 pixels# wh = wh * (npr.rand(wh.shape[0], 1) * 0.9 + 0.1) # multiply by random scale 0-1# Kmeans inittry:LOGGER.info(f"{PREFIX}Running kmeans for {n} anchors on {len(wh)} points...")assert n <= len(wh) # apply overdetermined constraints = wh.std(0) # sigmas for whiteningk = kmeans(wh / s, n, iter=30)[0] * s # pointsassert n == len(k) # kmeans may return fewer points than requested if wh is insufficient or too similarexcept Exception:LOGGER.warning(f"{PREFIX}WARNING ⚠️ switching strategies from kmeans to random init")k = np.sort(npr.rand(n * 2)).reshape(n, 2) * img_size # random initwh, wh0 = (torch.tensor(x, dtype=torch.float32) for x in (wh, wh0))k = print_results(k, verbose=False)# Plot# k, d = [None] * 20, [None] * 20# for i in tqdm(range(1, 21)):# k[i-1], d[i-1] = kmeans(wh / s, i) # points, mean distance# fig, ax = plt.subplots(1, 2, figsize=(14, 7), tight_layout=True)# ax = ax.ravel()# ax[0].plot(np.arange(1, 21), np.array(d) ** 2, marker='.')# fig, ax = plt.subplots(1, 2, figsize=(14, 7)) # plot wh# ax[0].hist(wh[wh[:, 0]<100, 0],400)# ax[1].hist(wh[wh[:, 1]<100, 1],400)# fig.savefig('wh.png', dpi=200)# Evolvef, sh, mp, s = anchor_fitness(k), k.shape, 0.9, 0.1 # fitness, generations, mutation prob, sigmapbar = tqdm(range(gen), bar_format=TQDM_BAR_FORMAT) # progress barfor _ in pbar:v = np.ones(sh)while (v == 1).all(): # mutate until a change occurs (prevent duplicates)v = ((npr.random(sh) < mp) * random.random() * npr.randn(*sh) * s + 1).clip(0.3, 3.0)kg = (k.copy() * v).clip(min=2.0)fg = anchor_fitness(kg)if fg > f:f, k = fg, kg.copy()pbar.desc = f"{PREFIX}Evolving anchors with Genetic Algorithm: fitness = {f:.4f}"if verbose:print_results(k, verbose)return print_results(k).astype(np.float32)
- 核查的主要代码:
@TryExcept(f"{PREFIX}ERROR")
def check_anchors(dataset, model, thr=4.0, imgsz=640):"""Evaluates anchor fit to dataset and adjusts if necessary, supporting customizable threshold and image size."""m = model.module.model[-1] if hasattr(model, "module") else model.model[-1] # Detect()shapes = imgsz * dataset.shapes / dataset.shapes.max(1, keepdims=True)scale = np.random.uniform(0.9, 1.1, size=(shapes.shape[0], 1)) # augment scalewh = torch.tensor(np.concatenate([l[:, 3:5] * s for s, l in zip(shapes * scale, dataset.labels)])).float() # whdef metric(k): # compute metricr = wh[:, None] / k[None]x = torch.min(r, 1 / r).min(2)[0] # ratio metricbest = x.max(1)[0] # best_xaat = (x > 1 / thr).float().sum(1).mean() # anchors above thresholdbpr = (best > 1 / thr).float().mean() # best possible recallreturn bpr, aatstride = m.stride.to(m.anchors.device).view(-1, 1, 1) # model stridesanchors = m.anchors.clone() * stride # current anchorsbpr, aat = metric(anchors.cpu().view(-1, 2))s = f"\n{PREFIX}{aat:.2f} anchors/target, {bpr:.3f} Best Possible Recall (BPR). "if bpr > 0.98: # threshold to recomputeLOGGER.info(f"{s}Current anchors are a good fit to dataset ✅")else:LOGGER.info(f"{s}Anchors are a poor fit to dataset ⚠️, attempting to improve...")na = m.anchors.numel() // 2 # number of anchorsanchors = kmean_anchors(dataset, n=na, img_size=imgsz, thr=thr, gen=1000, verbose=False)new_bpr = metric(anchors)[0]if new_bpr > bpr: # replace anchorsanchors = torch.tensor(anchors, device=m.anchors.device).type_as(m.anchors)m.anchors[:] = anchors.clone().view_as(m.anchors)check_anchor_order(m) # must be in pixel-space (not grid-space)m.anchors /= strides = f"{PREFIX}Done ✅ (optional: update model *.yaml to use these anchors in the future)"else:s = f"{PREFIX}Done ⚠️ (original anchors better than new anchors, proceeding with original anchors)"LOGGER.info(s)ps:
bpr(best possible recall)
aat(anchors above threshold)其中 bpr 参数就是判断是否需要重新计算锚定框的依据(是否小于0.98)。
- 重新计算符合此数据集标注框的锚定框,是利用
kmean聚类方法实现的,主要代码如下:
def kmean_anchors(dataset="./data/coco128.yaml", n=9, img_size=640, thr=4.0, gen=1000, verbose=True):"""Creates kmeans-evolved anchors from training dataset.Arguments:dataset: path to data.yaml, or a loaded datasetn: number of anchorsimg_size: image size used for trainingthr: anchor-label wh ratio threshold hyperparameter hyp['anchor_t'] used for training, default=4.0gen: generations to evolve anchors using genetic algorithmverbose: print all resultsReturn:k: kmeans evolved anchorsUsage:from utils.autoanchor import *; _ = kmean_anchors()"""from scipy.cluster.vq import kmeansnpr = np.randomthr = 1 / thrdef metric(k, wh): # compute metricsr = wh[:, None] / k[None]x = torch.min(r, 1 / r).min(2)[0] # ratio metric# x = wh_iou(wh, torch.tensor(k)) # iou metricreturn x, x.max(1)[0] # x, best_xdef anchor_fitness(k): # mutation fitness_, best = metric(torch.tensor(k, dtype=torch.float32), wh)return (best * (best > thr).float()).mean() # fitnessdef print_results(k, verbose=True):k = k[np.argsort(k.prod(1))] # sort small to largex, best = metric(k, wh0)bpr, aat = (best > thr).float().mean(), (x > thr).float().mean() * n # best possible recall, anch > thrs = (f"{PREFIX}thr={thr:.2f}: {bpr:.4f} best possible recall, {aat:.2f} anchors past thr\n"f"{PREFIX}n={n}, img_size={img_size}, metric_all={x.mean():.3f}/{best.mean():.3f}-mean/best, "f"past_thr={x[x > thr].mean():.3f}-mean: ")for x in k:s += "%i,%i, " % (round(x[0]), round(x[1]))if verbose:LOGGER.info(s[:-2])return kif isinstance(dataset, str): # *.yaml filewith open(dataset, errors="ignore") as f:data_dict = yaml.safe_load(f) # model dictfrom utils.dataloaders import LoadImagesAndLabelsdataset = LoadImagesAndLabels(data_dict["train"], augment=True, rect=True)# Get label whshapes = img_size * dataset.shapes / dataset.shapes.max(1, keepdims=True)wh0 = np.concatenate([l[:, 3:5] * s for s, l in zip(shapes, dataset.labels)]) # wh# Filteri = (wh0 < 3.0).any(1).sum()if i:LOGGER.info(f"{PREFIX}WARNING ⚠️ Extremely small objects found: {i} of {len(wh0)} labels are <3 pixels in size")wh = wh0[(wh0 >= 2.0).any(1)].astype(np.float32) # filter > 2 pixels# wh = wh * (npr.rand(wh.shape[0], 1) * 0.9 + 0.1) # multiply by random scale 0-1# Kmeans inittry:LOGGER.info(f"{PREFIX}Running kmeans for {n} anchors on {len(wh)} points...")assert n <= len(wh) # apply overdetermined constraints = wh.std(0) # sigmas for whiteningk = kmeans(wh / s, n, iter=30)[0] * s # pointsassert n == len(k) # kmeans may return fewer points than requested if wh is insufficient or too similarexcept Exception:LOGGER.warning(f"{PREFIX}WARNING ⚠️ switching strategies from kmeans to random init")k = np.sort(npr.rand(n * 2)).reshape(n, 2) * img_size # random initwh, wh0 = (torch.tensor(x, dtype=torch.float32) for x in (wh, wh0))k = print_results(k, verbose=False)# Plot# k, d = [None] * 20, [None] * 20# for i in tqdm(range(1, 21)):# k[i-1], d[i-1] = kmeans(wh / s, i) # points, mean distance# fig, ax = plt.subplots(1, 2, figsize=(14, 7), tight_layout=True)# ax = ax.ravel()# ax[0].plot(np.arange(1, 21), np.array(d) ** 2, marker='.')# fig, ax = plt.subplots(1, 2, figsize=(14, 7)) # plot wh# ax[0].hist(wh[wh[:, 0]<100, 0],400)# ax[1].hist(wh[wh[:, 1]<100, 1],400)# fig.savefig('wh.png', dpi=200)# Evolvef, sh, mp, s = anchor_fitness(k), k.shape, 0.9, 0.1 # fitness, generations, mutation prob, sigmapbar = tqdm(range(gen), bar_format=TQDM_BAR_FORMAT) # progress barfor _ in pbar:v = np.ones(sh)while (v == 1).all(): # mutate until a change occurs (prevent duplicates)v = ((npr.random(sh) < mp) * random.random() * npr.randn(*sh) * s + 1).clip(0.3, 3.0)kg = (k.copy() * v).clip(min=2.0)fg = anchor_fitness(kg)if fg > f:f, k = fg, kg.copy()pbar.desc = f"{PREFIX}Evolving anchors with Genetic Algorithm: fitness = {f:.4f}"if verbose:print_results(k, verbose)return print_results(k).astype(np.float32)
参数释意:
- dataset:包含数据集文件路径等相关信息的 yaml 文件,或者数据集张量(yolov5 自动计算锚定框时就是用的这种方式,先把数据集标签信息读取再处理)。默认 coco128.yaml
- n:锚定框的数量,即有几组。默认值是9
- img_size:图像尺寸。计算数据集样本标签框的宽高比时,是需要缩放到 img_size 大小后再计算的。默认值是640
- thr:数据集中标注框宽高比最大阈值,默认使用超参文件./data/hyps/hyp.scratch- .yaml 中的 “anchor_t”参数值;默认值是4.0。自动计算时,会自动根据你所使用的数据集,来计算合适的阈值。
- gen:kmean聚类算法迭代次数。默认值是1000
- verbose:是否打印输出所有计算结果,默认值是true
- 如果不想自动计算锚定框,可以在train.py中设置参数:
parser.add_argument('--noautoanchor', action='store_true', help='disable autoanchor check')3、手动锚框计算
- 1. 在./data文件夹下复制VOC.yaml文件,自己命名,如train_data.yaml文件,修改文件路径为绝对路径
train: # train images (relative to 'path') 16551 imagesF:/dataset/yolo/yolov5_up_sum/yolov5-master/datasets/train_data/images/trainval: # val images (relative to 'path') 4952 imagesF:/dataset/yolo/yolov5_up_sum/yolov5-master/datasets/train_data/images/val
test: # test images (optional)# Classes
names: ['Team1', 'Team2', 'Ball', 'Team3']- 数据集中需含有.cache文件

如果数据集中不存在.cache文件,查找Yolov5训练自己数据集的帖子,按照流程运行train.py文件,成功的话文件夹下会自动生成.cache文件
.cache文件:原始数据里没有该文件,yolov5自动生成的缓存文件,再下次读数据时,直接读取缓存文件,速度更快
- 2. 在Yolov5目录下新建一个.py文件,调用kmeans算法计算anchor:
import utils.autoanchor as autoACif __name__ == '__main__':config = "./data/train_data.yaml"# 对数据集重新计算 anchorsnew_anchors = autoAC.kmean_anchors(config, 9, 640, 5.0, 1000, True)print(new_anchors)
运行结果展示:
albumentations: Blur(p=0.01, blur_limit=(3, 7)), MedianBlur(p=0.01, blur_limit=(3, 7)), ToGray(p=0.01), CLAHE(p=0.01, clip_limit=(1, 4.0), tile_grid_size=(8, 8))
Scanning F:\dataset\yolo\yolov5_up_sum\yolov5-master\datasets\train_data\labels\train... 107 images, 0 backgrounds, 0 corrupt: 100%|██████████| 107/107 [00:15<00:00, 6.94it/s]
WARNING Cache directory F:\dataset\yolo\yolov5_up_sum\yolov5-master\datasets\train_data\labels is not writeable: [WinError 183] : 'F:\\dataset\\yolo\\yolov5_up_sum\\yolov5-master\\datasets\\train_data\\labels\\train.cache.npy' -> 'F:\\dataset\\yolo\\yolov5_up_sum\\yolov5-master\\datasets\\train_data\\labels\\train.cache'
AutoAnchor: Running kmeans for 9 anchors on 855 points...0%| | 0/1000 [00:00<?, ?it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.79 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.399/0.789-mean/best, past_thr=0.488-mean: 15,20, 25,23, 20,46, 35,39, 34,71, 62,60, 76,129, 100,213, 162,232
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.83 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.400/0.789-mean/best, past_thr=0.486-mean: 15,20, 25,23, 20,47, 36,40, 34,67, 63,62, 74,126, 102,216, 159,232
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.81 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.399/0.790-mean/best, past_thr=0.485-mean: 15,19, 25,23, 20,47, 36,39, 34,69, 63,62, 73,128, 97,212, 161,230
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.82 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.398/0.790-mean/best, past_thr=0.483-mean: 15,19, 25,23, 19,46, 36,39, 32,67, 64,63, 74,131, 101,205, 161,220
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.398/0.790-mean/best, past_thr=0.483-mean: 15,19, 25,23, 20,46, 35,39, 33,67, 64,63, 74,131, 101,205, 160,219
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.399/0.791-mean/best, past_thr=0.484-mean: 15,19, 25,23, 19,46, 35,39, 32,66, 64,65, 74,131, 100,205, 160,222
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.399/0.791-mean/best, past_thr=0.484-mean: 15,19, 25,23, 20,46, 35,39, 32,66, 64,64, 74,131, 100,205, 160,221
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.88 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.792-mean/best, past_thr=0.492-mean: 14,20, 25,24, 19,45, 34,38, 33,59, 60,64, 73,128, 97,209, 147,221
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.88 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.408/0.794-mean/best, past_thr=0.494-mean: 14,20, 25,22, 20,45, 30,36, 33,58, 60,59, 73,123, 90,203, 153,229
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.88 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.408/0.794-mean/best, past_thr=0.494-mean: 14,20, 25,22, 20,45, 30,36, 33,58, 60,59, 74,124, 89,204, 154,228
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.89 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.408/0.795-mean/best, past_thr=0.493-mean: 14,19, 25,22, 20,44, 31,36, 34,57, 60,59, 75,121, 89,211, 154,225
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.89 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.407/0.795-mean/best, past_thr=0.493-mean: 14,19, 25,22, 20,44, 31,36, 34,57, 59,59, 75,121, 89,212, 154,225
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.88 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.407/0.795-mean/best, past_thr=0.493-mean: 14,19, 25,22, 20,44, 31,36, 34,57, 59,59, 75,122, 89,213, 155,224
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.403/0.795-mean/best, past_thr=0.489-mean: 14,19, 24,21, 19,44, 31,37, 35,57, 60,49, 76,122, 93,226, 146,224
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.82 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.404/0.795-mean/best, past_thr=0.491-mean: 14,19, 24,21, 19,44, 31,37, 35,58, 60,50, 76,121, 94,227, 148,224
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.82 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.404/0.796-mean/best, past_thr=0.491-mean: 14,19, 24,21, 19,44, 31,37, 35,58, 59,50, 76,121, 94,228, 149,225
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.404/0.796-mean/best, past_thr=0.491-mean: 14,19, 25,21, 19,44, 30,36, 33,58, 60,52, 75,116, 92,228, 149,227
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7959: 16%|█▌ | 156/1000 [00:00<00:00, 1559.97it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.404/0.796-mean/best, past_thr=0.492-mean: 14,19, 25,21, 19,44, 30,36, 33,58, 60,52, 75,117, 92,228, 148,227
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.404/0.796-mean/best, past_thr=0.490-mean: 14,18, 24,20, 18,44, 29,36, 33,58, 59,53, 73,116, 91,228, 149,225
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.88 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.490-mean: 14,18, 23,20, 19,44, 29,36, 33,58, 59,53, 74,118, 92,219, 150,226
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 23,20, 19,43, 30,36, 32,58, 59,52, 73,119, 92,220, 148,230
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 23,20, 19,43, 30,36, 33,58, 59,53, 73,122, 92,220, 148,228
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 23,20, 19,43, 30,37, 33,58, 59,52, 72,122, 93,220, 147,229
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 23,20, 19,43, 30,37, 33,58, 59,52, 72,122, 93,220, 147,229
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 23,20, 19,43, 30,36, 33,58, 59,52, 73,122, 93,220, 146,230
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 24,20, 19,43, 30,36, 33,58, 58,53, 73,122, 93,222, 147,227
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7964: 32%|███▏ | 321/1000 [00:00<00:00, 1600.54it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 24,20, 19,43, 30,37, 33,58, 58,53, 73,122, 93,222, 147,227
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 24,20, 19,43, 30,36, 33,58, 58,53, 73,122, 93,221, 147,227
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 24,20, 19,43, 30,36, 33,58, 58,53, 73,122, 93,222, 147,227
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.796-mean/best, past_thr=0.491-mean: 14,18, 24,20, 19,43, 30,36, 33,58, 58,53, 73,123, 92,222, 147,227
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,20, 19,42, 30,36, 33,57, 58,54, 72,122, 93,218, 147,230
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,20, 19,42, 30,35, 33,57, 57,54, 72,122, 93,218, 146,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.407/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,20, 19,43, 30,36, 32,57, 57,54, 72,122, 93,218, 143,234
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,21, 19,42, 30,36, 32,57, 58,55, 72,123, 93,219, 143,234
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7968: 48%|████▊ | 482/1000 [00:00<00:00, 1548.48it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,21, 19,42, 30,36, 32,58, 58,55, 72,123, 92,220, 143,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,19, 24,21, 19,42, 31,36, 32,58, 59,55, 72,122, 93,218, 143,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,19, 24,21, 19,42, 31,36, 32,58, 59,55, 72,122, 93,218, 143,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.87 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,21, 19,42, 31,37, 32,58, 59,56, 73,123, 93,218, 143,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,21, 19,42, 31,37, 32,58, 59,56, 72,123, 92,218, 143,235
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,21, 19,42, 31,36, 32,59, 57,57, 72,126, 92,217, 142,240
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.84 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.493-mean: 14,18, 24,21, 19,42, 31,36, 32,59, 58,57, 72,126, 91,217, 142,240
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7971: 65%|██████▍ | 647/1000 [00:00<00:00, 1563.54it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,21, 19,42, 31,36, 32,59, 57,57, 73,126, 92,216, 142,239
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7971: 82%|████████▏ | 816/1000 [00:00<00:00, 1607.99it/s]AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,20, 19,42, 31,36, 32,58, 57,57, 72,126, 92,215, 140,237
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.86 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.406/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,20, 19,42, 31,36, 32,58, 57,57, 71,126, 92,214, 140,236
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,20, 19,42, 31,36, 32,58, 57,57, 71,126, 92,214, 140,236
[[ 14.077 18.166][ 23.833 20.45][ 18.59 42.161][ 30.656 36.243][ 32.122 58.356][ 57.303 56.757][ 71.018 126.44][ 91.503 214.03][ 140.23 235.73]]
AutoAnchor: Evolving anchors with Genetic Algorithm: fitness = 0.7972: 100%|██████████| 1000/1000 [00:00<00:00, 1630.48it/s]
AutoAnchor: thr=0.20: 1.0000 best possible recall, 6.85 anchors past thr
AutoAnchor: n=9, img_size=640, metric_all=0.405/0.797-mean/best, past_thr=0.492-mean: 14,18, 24,20, 19,42, 31,36, 32,58, 57,57, 71,126, 92,214, 140,236Process finished with exit code 0
输出的9个坐标即为锚框中心坐标,复制yolov5s.yaml文件,自己命名,如yolov5s_train.yaml,将计算所得值按顺序修改至模型配置文件./model/yolov5s_train.yaml中,重新训练即可:
# YOLOv5 🚀 by Ultralytics, AGPL-3.0 license# Parameters
nc: 4 # number of classes
depth_multiple: 0.33 # model depth multiple
width_multiple: 0.50 # layer channel multiple
anchors:- [14.077, 18.166, 23.833, 20.45, 18.59, 42.161] # P3/8- [30.656, 36.243, 32.122, 58.356, 57.303, 56.757] # P4/16- [71.018, 126.44, 91.503, 214.03, 140.23, 235.73] # P5/324. 检测模块
(没看太懂,后面再查些资料)
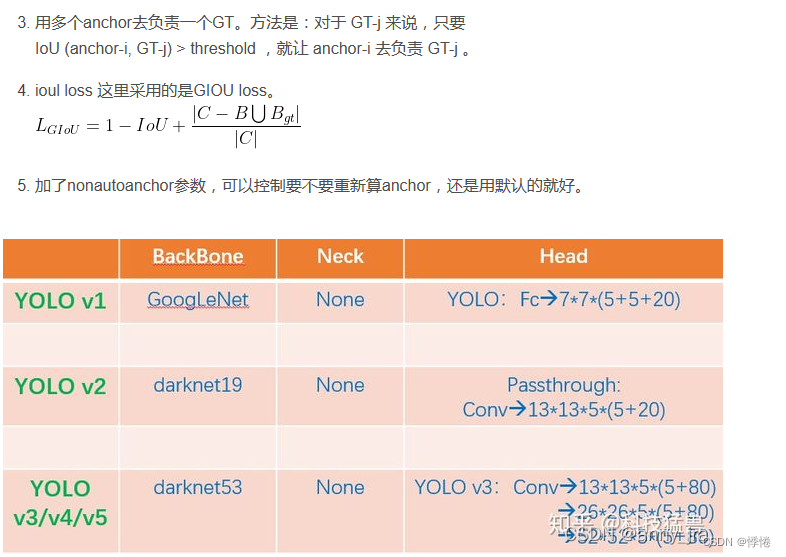
anchor在模型中的应用涉及到了yolo系列目标框回归的过程。yolov5中的detect模块沿用了v3检测方式。
- 1. 检测到的不是框而是偏移量: tx,ty指的是针对所在grid的左上角坐标的偏移量, tw,th指的是相对于anchor的宽高的偏移量,通过如下图的计算方式,得到bx,by,bw,bh就是最终的检测结果。

- 2. 前面经过backbone,neck,head是panet的三个分支,可见特征图size不同,每个特征图分了13个网格,同一尺度的特征图对应了3个anchor,检测了[c,x,y,w,h]和num_class个的one-hot类别标签。3个尺度的特征图,总共就有9个anchor。

参考:
Yolov5的anchors设置详解
Yolov5的anchor详解
YOLOv5的anchor设定
(20)目标检测算法之YOLOv5计算预选框、详解anchor计算